d1b05a_d1lc0a2
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1b05_Ah
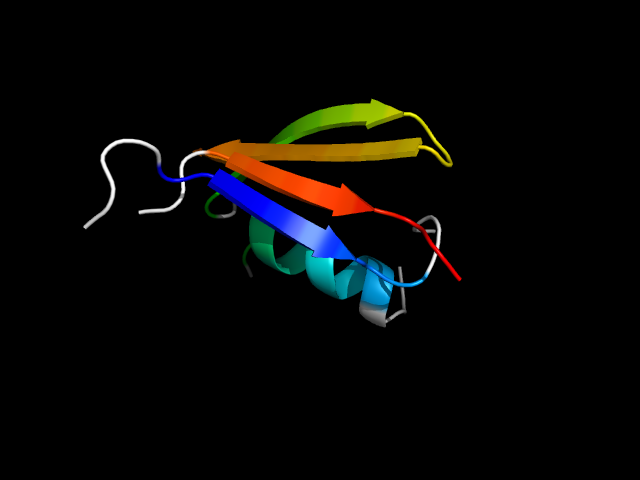 |
d1b05a_ | A9-A24,A31-A46,A188-A207,A217-A227 | Alpha and beta proteins (a/b) | Periplasmic binding protein-like II | Periplasmic binding protein-like II | Phosphate binding protein-like | Oligo-peptide binding protein (OPPA) |
1lc0_Ac
 |
d1lc0a2 | A129-A159,A160-A180,A181-A207,A208-A246 | Alpha and beta proteins (a+b) | Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain | Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain | Biliverdin reductase | Biliverdin reductase
|
Justification for analogy
The motif in 1b05: a hybrid motif, because a beta-hairpin is absent in the other duplicate and thus insertion.
The motif in 1lc0: although "Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain" is a very variable fold, it is in SCOP a+b class and
does not have any resemblance to Rossmann-like structures. 1b05 is in a/b class and is Rossmann-like.
So these two motifs are analogs.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b05_Ah ------------ladkQTLVRNNGsevq-------iegvPESNVSRDLFEg-ngAYKLKNWVVN---------ERIVLERNvinQVTYLPIS----------------------------
1lc0_Ac meefeflrrevlgkelLKGSLRFTaspleeerfgfpafsGISRLTWLVSLfg--ELSLISATLEerkedqymkMTVQLETQnkgLLSWIEEKgpglkrnryvnfqftsgsleevpsvgvn
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b05_Ah ------------LADKQTLVRNNG----------sEVQIegvPESNVSRDLFegnGAYKLKNWVV---------NERIVLERNVINQVTYLPIS----------------------------
1lc0_Ac meefeflrrevlGKELLKGSLRFTaspleeerfgfPAFS---GISRLTWLVS-lfGELSLISATLeerkedqymKMTVQLETQNKGLLSWIEEKgpglkrnryvnfqftsgsleevpsvgvn
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b05_Ah -------------LADKQTLVRNNGS---------EVQIEGVPESNVSR--D---LFEGNGAYKLKNWVV---------NERIVLERNVINQVTYLPIS----------------------------
1lc0_Ac MEEFEFLRREVLGKE-LLKGSLRFTASPLEEERFGFPAF-S-GISRL-TWLVSLF-----GELSLISATLEERKEDQYMKMTVQLETQNKGLLSWIEEKGPGLKRNRYVNFQFTSGSLEEVPSVGVN
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b05_Ah LADKQ-----------------TLVRNNGSEVQIEG------------------VPESNVSRDLFEGNGA--YKLKNWVVN----------ERIVLERNVIN--QVTYLPIS----------------------------
1lc0_Ac -----MEEFEFLRREVLGKELLKGSLRF--------TASPLEEERFGFPAFSGISRLTWLVSLF------GELSLISATL-EERKEDQYMKMTVQLETQN--KGLLSWIEEKGPGLKRNRYVNFQFTSGSLEEVPSVGVN

