d1b05a_d1nkia_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1b05_Ah
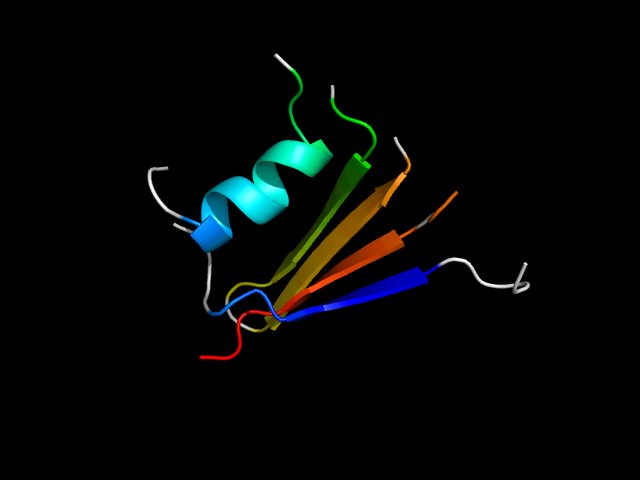 |
d1b05a_ | A9-A24,A31-A46,A188-A207,A217-A227 | Alpha and beta proteins (a/b) | Periplasmic binding protein-like II | Periplasmic binding protein-like II | Phosphate binding protein-like | Oligo-peptide binding protein (OPPA) |
1nki_Ac
 |
d1nkia_ | A1-A13,A14-A25,A26-A43,A44-A57 | Alpha and beta proteins (a+b) | Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase | Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase | Antibiotic resistance proteins | Fosfomycin resistance protein A (FosA)
|
Justification for analogy
The babbb motif in 1b05: a hybrid motif, because a beta-hairpin is absent in the majority of homologs in this superfamily and therefore is an insertion. Other parts of the motif come from the core of this superfamily.
The babbb motif in 1nki: a core motif.
Thus, these two motifs are analogs.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b05_Ah -ladkqTLVRNNGsevqiegVPESNVSRDLFEgnGAYKLKNWVVnERIVLERnVInQVTYLPIS----
1nki_Ac mltglnHLTLAVA-------DLPASIAFYRDL--LGFRLEARWD-QGAYLELgSL-WLCLSREPqygg
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b05_Ah -LADKQTLVRNNGSevqiegvPESNVSRDLFEgnGAYKLKNWVVnERIVLERnVINQVTYLPIS----
1nki_Ac mLTGLNHLTLAVAD-------LPASIAFYRDL--LGFRLEARWD-QGAYLEL-GSLWLCLSREPqygg
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b05_Ah --LADKQTLVRNNGSEVQIEGVPESNVSRDLFEGNGAYKLKNWVVNERIVLERNVINQVTYLPIS----
1nki_Ac ML-TGLNHLTLAVADLP------A-SIAFYRDL-LG-FRLEARWD-QGAYLELG-SLWLCLSREPQYGG
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b05_Ah LAD----KQTLVRNNGSEVQIEGVPESNVSRDLFEGN-GAYKLKNWVVNERIVLERNVIN----QVTYLPIS----
1nki_Ac ---MLTGLNHLTLAVAD-------LPASIAFYRD---LLGFRLEARWD-QGAYLE-----LGSLWLCLSREPQYGG

