d1b0pa2d1k0da2
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1b0p_Ah
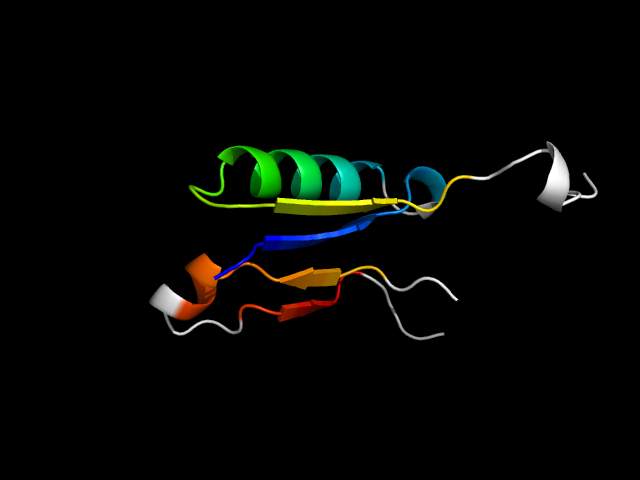 |
d1b0pa2 | A1034-A1078,A1095-A1120 | Alpha and beta proteins (a/b) | Thiamin diphosphate-binding fold (THDP-binding) | Thiamin diphosphate-binding fold (THDP-binding) | PFOR PP module | Pyruvate-ferredoxin oxidoreductase, PFOR, domains VI |
1k0d_Ac
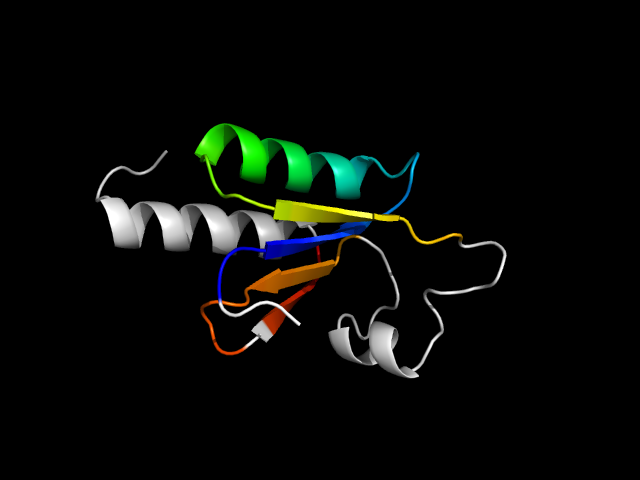 |
d1k0da2 | A109-A161,A162-A200 | Alpha and beta proteins (a/b) | Thioredoxin fold | Thioredoxin-like | Glutathione S-transferase (GST), N-terminal domain | Yeast prion protein ure2p, nitrogen regulation fragment
|
Justification for analogy
The motif in 1b0p: hybrid, since the C-terminal beta-hairpin is absent in d1b0pa1, which is the other module on the same chain and in the same superfamily.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b0p_Ah ---YVYVATVSMGysKQQFLKVLKEAESFPGPSLVIAYAtcinqglrk--------ywpLFRYDPRLAaqgknPFQLDskap----------------
1k0d_Ac qplEGYTLFSHRS--APNGFKVAIVLSELGFHYNTIFLDfnlgehrapefvsvnpnarvPALIDHGMD----nLSIWEsgaillhlvnkyyketgnpl
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b0p_Ah ---YVYVATVSMGysKQQFLKVLKEAESFPGPSLVIAYATC-------inqglrkYWPL-FRYDPRLAaqgknPFQLDS----------------kap
1k0d_Ac qplEGYTLFSHRS--APNGFKVAIVLSELGFHYNTIFLDFNlgehrapefvsvnpNARVpALIDHGMD----nLSIWESgaillhlvnkyyketgnpl
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b0p_Ah ---YVYVATVS-MGYSKQQFLKVLKEAESFPGPSLVIAYAT-CINQ-GLRKYW-----------P-L-FRYDPRLA-AQGKNPFQLD--------------------SKAP
1k0d_Ac QPLEGYTLFSHRS--AP-NGFKVAIVLSELGFHYNTIFLDFN---LG----EHRAPEFVSVNPNARVPALIDHG-MD----NLSIWESGAILLHLVNKYYKETGNPL----
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b0p_Ah ---YVYVATVSMGYSKQQFLKVLKEAESFP-GPSLVIAYATCINQGLRKYWP-------------------LFRYDPRLAAQGKNPFQLDSKAP---------------------------
1k0d_Ac QPLEGYTLFSHRS-APNGFKVAIVLSEL--GFHYNTIFLDF-----------NLGEHRAPEFVSVNPNARVPALIDHGM---------------DNLSIWESGAILLHLVNKYYKETGNPL

