d1b6ra1d1v97a4
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1b6r_Ai
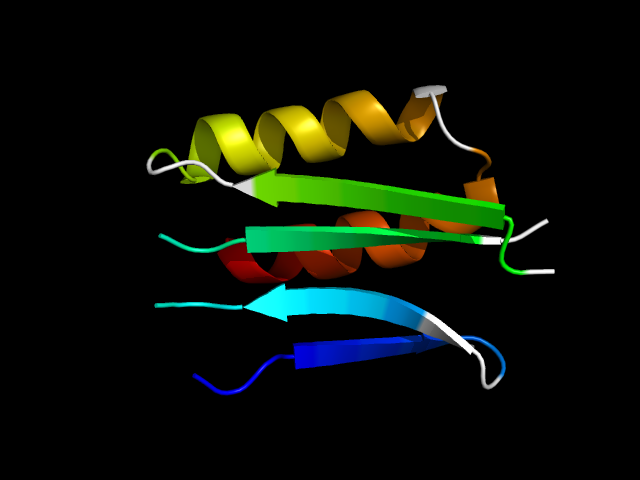 |
d1b6ra1 | A178-A196,A278-A287,A313-A355 | All beta proteins | Barrel-sandwich hybrid | Rudiment single hybrid motif | BC C-terminal domain-like | N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), C-domain |
1v97_Ac
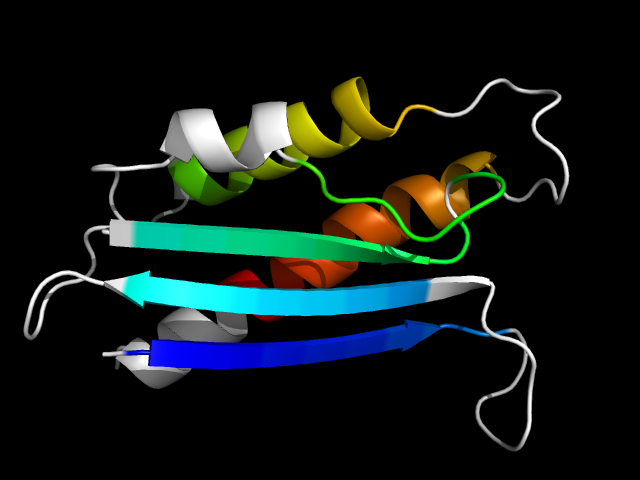 |
d1v97a4 | A415-A444,A445-A459,A460-A528 | Alpha and beta proteins (a+b) | CO dehydrogenase flavoprotein C-domain-like | CO dehydrogenase flavoprotein C-terminal domain-like | CO dehydrogenase flavoprotein C-terminal domain-like | Xanthine oxidase, domain 4 (?)
|
Justification for analogy
The motif in 1b6r: motif occurs at the interface of the ATP-grasp fold and 'Barrel-sandwich hybrid' fold.
The motif in 1v97: stand-along, core motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b6r_Ai -PLTHNLHQD-------gilRTSVAFP---------NPSVMINLigrKVGHLNLtds------------DTSRLTATLEALIPll-------PPEYASGVIWAQSKFG--------
1v97_Ac dEFFSAFKQAsrreddiakvTCGMRVLfqpgsmqvkELALCYGGm--ADRTISAlkttqkqlskfwnekLLQDVCAGLAEELSlspdapggmIEFRRTLTLSFFFKFYltvlkklg
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b6r_Ai -PLTHNLHQD-------GILRTSVAFP---------NPSVMINLigrKVGHLNL---TDSD---------TSRLTATLEALIPL-------lPPEY-ASGVIWAQSKFG-------
1v97_Ac dEFFSAFKQAsrreddiAKVTCGMRVLfqpgsmqvkELALCYGG--mADRTISAlktTQKQlskfwneklLQDVCAGLAEELSLspdapggmIEFRrTLTLSFFFKFYLtvlkklg
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b6r_Ai PLTHNLHQD------------GILRTSVAFP-N-PS-VMINLI--G---RKVGH--LN-L-TDSDTSRLTATLEALIPL-L----------------------PPEYASGVIWAQS-KFG
1v97_Ac -DEFFSAFKQASRREDDIAKVTCGMRVLFQPGSMQVKELALCYGGMADRTISALKTTQKQLSK--F--WNEKLLQDVCAGLAEELSLSPDAPGGMIEFRRTLTLSFFFKFYLTVLKKL-G
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1b6r_Ai -PLTHNLHQDGILR------------TSVAFPN----------PSVMINLIGRKV----GHLNLTDSDT------------SRLTATLEA-LIPLLP---------------PEYASGVIWAQSKFG----
1v97_Ac DEFFSAFKQ-----ASRREDDIAKVTCGMRVL-FQPGSMQVKELALCYG------GMADRTISAL----KTTQKQLSKFWNEKLLQDVCAGLAEEL-SLSPDAPGGMIEFRRTLTLSFFFKFYLTVLKKLG

