d1ce8a6d1di2a_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1ce8_Ah
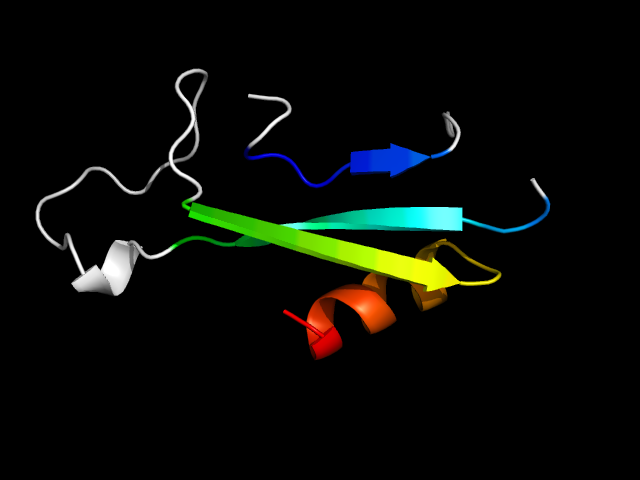 |
d1ce8a6 | A243-A255,A352-A402 | Alpha and beta proteins (a+b) | ATP-grasp | Glutathione synthetase ATP-binding domain-like | BC ATP-binding domain-like | Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains |
1di2_Ac
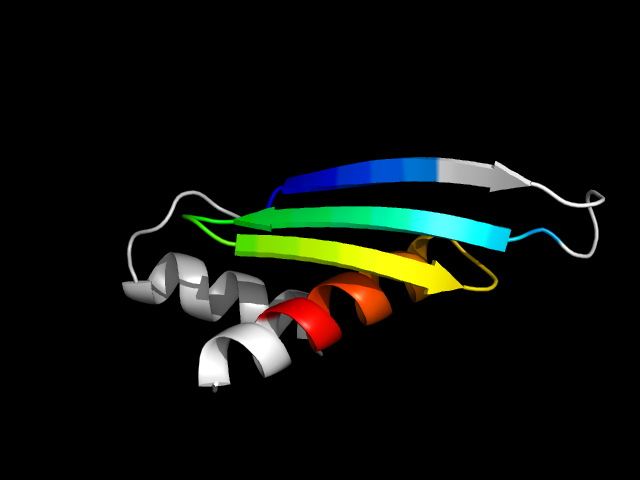 |
d1di2a_ | A112-A140,A141-A180 | Alpha and beta proteins (a+b) | dsRBD-like | dsRNA-binding domain-like | Double-stranded RNA-binding domain (dsRBD) | Double-stranded RNA-binding protein A, second dsRBD
|
Justification for analogy
The motif in 1ce8 uses one strand from the ATP-grasp core and two strands and one helix from C-terminal extension. Comparing d1ce8a6 with d1gsa_2
(different family same superfamily), the C-terminal extension is not present in d1gsa_2. So this is a hybrid motif. And it is analogous to real
dsRBD domains.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ce8_Ah --------------htgDSITVApaqt---iDYVVTKIPRFnfekfagandrlttqmksVGEVMAIGRTQQESLQKALRGL------
1di2_Ac mpvgslqelavqkgwrlPEYTVAqesgpphkREFTITCRVE------------------TFVETGSGTSKQVAKRVAAEKLltkfkt
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ce8_Ah --------------htgDSITVAP---aqtIDYVVTKIPRFnfekfagandrlttqmksVGEVMAIGRTQQESLQKALRGL------
1di2_Ac mpvgslqelavqkgwrlPEYTVAQesgpphKREFTITCRVE------------------TFVETGSGTSKQVAKRVAAEKLltkfkt
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ce8_Ah -----------------HTGDSITVAPAQTID-----YVVTKIPRFNFEKFAGANDRLTTQMKSVGEVMAIGRTQQESLQKALRGL------
1di2_Ac MPVGSLQELAVQKGWRL---PEYTVAQE--SGPPHKREFTITCRVE------------------TFVETGSGTSKQVAKRVAAEKLLTKFKT
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ce8_Ah HTGD------------------SITVAPAQTID--------YVVTKIPRFNFEKFAGANDRLTTQMKS-VGEVMAIGRTQQESLQKALRGL-------
1di2_Ac ----MPVGSLQELAVQKGWRLPEYTVAQ-----ESGPPHKREFTITCRV-------------------ETFVETGSGTSKQVAKRVAAEK-LLTKFKT

