d1cs1a_d1q8ia1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1cs1_Ah
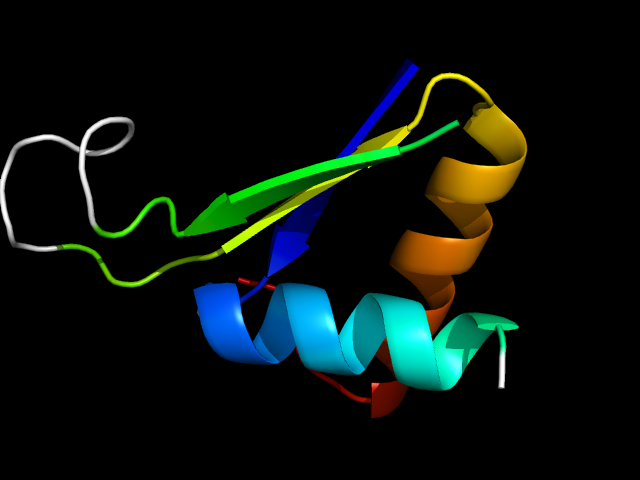 |
d1cs1a_ | A68-A88,A190-A233 | Alpha and beta proteins (a/b) | PLP-dependent transferases | PLP-dependent transferases | Cystathionine synthase-like | Cystathionine gamma-synthase, CGS |
1q8i_Ac
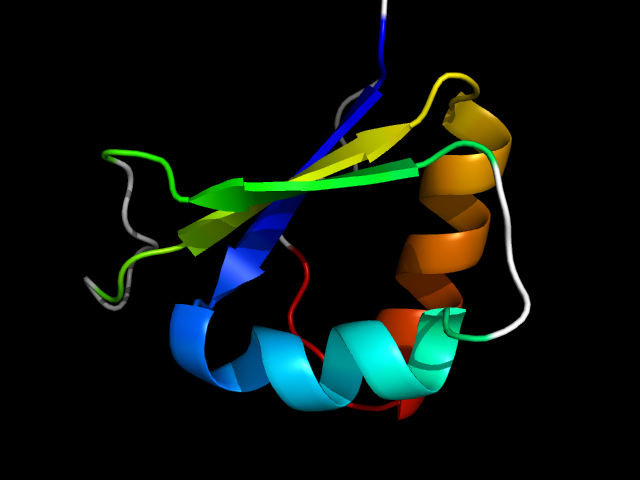 |
d1q8ia1 | A39-A60,A61-A109 | Alpha and beta proteins (a/b) | Ribonuclease H-like motif | Ribonuclease H-like | DnaQ-like 3'-5' exonuclease | Exonuclease domain of family B DNA polymerases
|
Justification for analogy
The motif in 1cs1: Although each family in the PLP-dependent transferases superfamily has this motif, this is a hybrid, because PLP-dependent
transferases superfamily is homologous to some flavodoxin proteins. So this motif is a hybrid between the Rossmann-like core and the insertions, and
is therefore analogous to ancient folds like ferredoxin.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1cs1_Ah -AGAVLTNtGMSAIHLVTTVFl--DLVLHSCTkylnghsdVVAGVVIAKDPDVVTELAWWANNIGVTG------
1q8i_Ac qESVAFIP-ADQVPRAQHILQgeqGFRLTPLAlkd--fhrQPVYGLYCRAHRQLMNYEKRLREGGVTVyeadvr
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1cs1_Ah -AGAVLTNTgMSAIHLVTTVFL--DLVLHSCTKylngHSDVVAGVVIAKDPDVVTELAWWANNIGVT------g
1q8i_Ac qESVAFIPA-DQVPRAQHILQGeqGFRLTPLAL--kdFHRQPVYGLYCRAHRQLMNYEKRLREGGVTvyeadvr
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1cs1_Ah -AGAVLTNTGMSAIHLVTTVFL--DLVLHSCTKYLN----GHSDVVAGVVIAKDPDVVTELAWWANNIGVTG------
1q8i_Ac QESVAFIPADQV-PRAQHILQGEQGFRLTPL---ALKDFH---RQPVYGLYCRAHRQLMNYEKRLREGGVTVYEADVR
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1cs1_Ah -AGAVLTNTGMSAIHLVTTVFL----DLVLHSCTKYLNG----HSDVVAGVVIAKDPDVVTELAWWANNIGVTG--------
1q8i_Ac QESVAFIP-ADQVPRAQHIL--QGEQGFRLTPL------ALKDFHRQPVYGLYCRAHRQLMNYEKRLREGGV--TVYEADVR

