d1dc1a_d1nf2a_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1dc1_Ah
 |
d1dc1a_ | A135-A206 | Alpha and beta proteins (a/b) | Restriction endonuclease-like | Restriction endonuclease-like | Restriction endonuclease BsobI | Restriction endonuclease BsobI |
1nf2_Ac
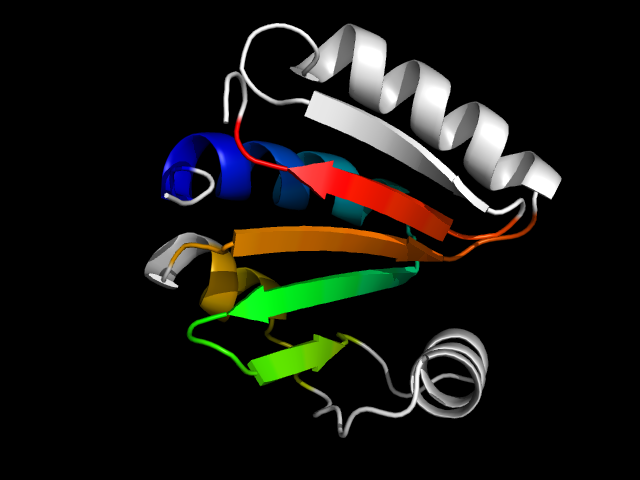 |
d1nf2a_ | A81-A189 | Alpha and beta proteins (a/b) | HAD-like | HAD-like | Predicted hydrolases Cof | Hypothetical protein TM0651
|
Justification for analogy
The motif in 1dc1: compared to d1ckqa_ (same superfamiliy), two strands are insertion;
The motif in 1nf2: the inserted domain appears like a circular-permuted ferredoxin, and most of the elements in the motif come from this
ferredoxin-like part.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dc1_Ah galaQQKLTRAIISALDIANIPYKWLDSrdkkyTNWMDK-------------------PEDDYELE--tFAKGISWTIN-------------------------gKHRTLMYnitvsl
1nf2_Ac -ekiPPEVAKDIIEYIKPLNVHWQAYID-----DVLYSEkdneeiksyarhsnvdyrvEPNLSELVskmGTTKLLLIDTperldelkeilserfkdvvkvfksfpTYLEIVPknv---
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dc1_Ah galaQQKLTRAIISALDIANIPYKWLDSrdkkyTNWMDK-------------------PEDD-YELE-TFAKGISWTING-------------------------KHRTLMYNITvsl
1nf2_Ac -ekiPPEVAKDIIEYIKPLNVHWQAYID-----DVLYSEkdneeiksyarhsnvdyrvEPNLsELVSkMGTTKLLLIDTPerldelkeilserfkdvvkvfksfpTYLEIVPKNV---
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dc1_Ah -GALAQQKLTRAIISALDIANIPYKWLDSRDKKYTNWMDK-------------------PED-DYELE-TFAKGISWTI---NGK----------------------HRTLMYNITVSL
1nf2_Ac E-K-IPPEVAKDIIEYIKPLNVHWQAYI-DD----VLYSEKDNEEIKSYARHSNVDYRVEPNLSELVSKMGTTKLLLIDTPERLDELKEILSERFKDVVKVFKSFPTYLEIVPKNV---
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dc1_Ah GALAQ----QKLTRAIISALDIANIPYKWLDSRDKKY-TNWMDKPEDD-------------------------YELETF-AKGISWTING--------------------------KHRTLMYNITVSL--
1nf2_Ac -----EKIPPEVAKDIIEYIKPLNVHWQAYI------DDVLYS-----EKDNEEIKSYARHSNVDYRVEPNLSELVSKMGTTKLLLIDT-PERLDELKEILSERFKDVVKVFKSFPTYLEIVPK-----NV

