d1dmza_d1hdha_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1dmz_Ah
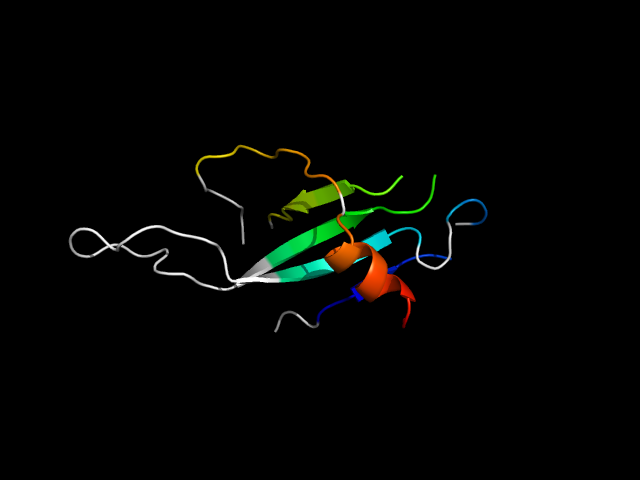 |
d1dmza_ | A596-A605,A615-A654,A666-A674,A706-A730 | All beta proteins | SMAD/FHA domain | SMAD/FHA domain | FHA domain | Phosphotyrosine binding domain of Rad53 |
1hdh_Ac
 |
d1hdha_ | A447-A462,A463-A479,A480-A493,A494-A527 | Alpha and beta proteins (a/b) | Alkaline phosphatase-like | Alkaline phosphatase-like | Arylsulfatase | Arylsulfatase B (4-sulfatase)
|
Justification for analogy
The motif in 1dmz: compare to d1gxca_ (same family), the helix is the insertion. Can be analog to similar motifs in SCOP a+b class.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dmz_Ah ------qgvNPFFIGRdNRLsrvHCFIFKkrhavgksmyespaqglddIWYCHTGT------QGTKFLLQdglgmlQEQRVVLK-----qTAEEKDLVKKL-------
1hdh_Ac eteaahdenTVTGWEL-FGM---RAIRQG-------------------DWKAVYLPapvgpaTWQLYDLA----rdPGEIHDLAdsqpgkLAELIEHWKRYvsetgvv
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dmz_Ah ------QGVNPFFIGRdnRLSRvhCFIFKkrhavgksmyespaqglddIWYCHTGT------QGTKFLLQDglgmlqeqRVVLK-----qTAEEKDLVKKL-------
1hdh_Ac eteaahDENTVTGWEL--FGMR--AIRQG-------------------DWKAVYLPapvgpaTWQLYDLAR----dpgeIHDLAdsqpgkLAELIEHWKRYvsetgvv
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dmz_Ah -----QG-VNPFFIGRDNRLSRVHCFIFKKRHAVGKSMYESPAQGLDDIWYCHTGT------QGTKFLLQDGLGM---LQEQRVVLK---Q--TAEEKDLVKKL-------
1hdh_Ac ETEAAHDENTVTGWEL--FGM--RAIRQG-------------------DWKAVYLPAPVGPATWQLYDLA---RDPGE----IHDLADSQPGKLAELIEHWKRYVSETGVV
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dmz_Ah Q-------GVNPFFIGRDNRLSRVHCFIFKKRHAVGKSMYESPAQGLDDI--WYCHTGT-------QGTKFLLQDGLGMLQEQRVVLKQT-----------------AEEKDLVKKL--------
1hdh_Ac -ETEAAHDENTVTGWEL-FGMR---AIRQ---------------------GDWKAVYL-PAPVGPATWQLYDLA----------------RDPGEIHDLADSQPGKLAELIEHWKR-YVSETGVV

