d1ds1a_d1tig__
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1ds1_Ah
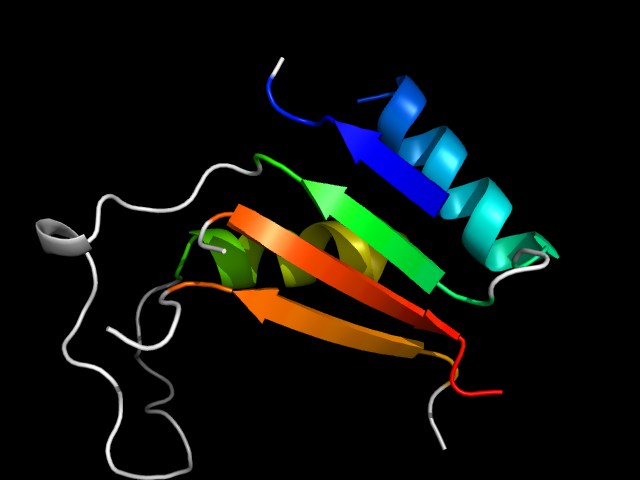 |
d1ds1a_ | A2-A8,A43-A105,A153-A166,A265-A275 | All beta proteins | Double-stranded beta-helix | Clavaminate synthase-like | Clavaminate synthase | Clavaminate synthase |
1tig__c
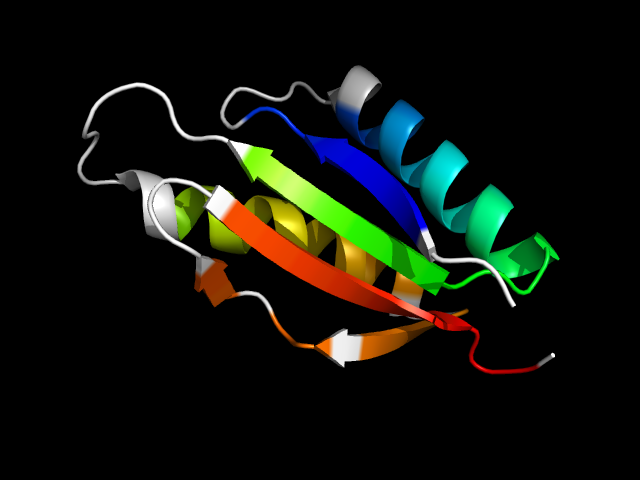 |
d1tig__ | 83-94,95-145,146-158,159-170 | Alpha and beta proteins (a+b) | IF3-like | Translation initiation factor IF3, C-terminal domain | Translation initiation factor IF3, C-terminal domain | Translation initiation factor IF3, C-terminal domain
|
Justification for analogy
Comparing d1ds1a_ and d1e5sa_ (same superfamily, different family), one strand and one helix are insertions.
Thus the IF3-like motif in d1ds1a_ is a hybrid.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ds1_Ah ---TSVDCTa------PGALATALDTFNAEGsedGHLLLRGlpveadadlpttpsstpapedrslLTMEAMLGLVGRRL-qpNYVM-LA-CSradhepGDLLIVDNF-
1tig__c invKEVRLSptieehdFNTKLRNARKFLEKG---DKVKATIrfkgr--------------aithkEIGQRVLDRLSEACa-dIAVVeTApKMdg--rnMFLVLAPKNd
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ds1_Ah ---TSVDCT------aPGALATALDTFNAEGSedgHLLLRgLPVEadADLPttpsstpapedrslLTMEAMLGLVGRRLqpNYVM--LACSRAdhePGDLLIVDNF-
1tig__c invKEVRLSptieehdFNTKLRNARKFLEKGD---KVKAT-IRFK--GRAI-----------thkEIGQRVLDRLSEACadIAVVetAPKMDG--rNMFLVLAPKNd
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ds1_Ah ---TSVDCT--A---P-GALATALDTFNAEGSEDGHLLLRGLPVEADADL-PTTPSSTPAPEDRSLLTMEAMLGLVGRRL---QPNYV-MLACSRADHEPGDLLIVDNF-
1tig__c INVKEVRLSPTIEEHDFNTKLRNARKFLEKGD---KVKATIRFKGR-AITH--------------KEIGQRVLDRLSEACADI--AVVETAPK-MDG-RNMFLVLAPKND
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ds1_Ah ---TSVDCTA-------PGALATALDTFNAEGSEDG-HLLLRGLPVEADADLPTTPSSTPAPEDRSLL------------TMEAMLGLVGRRLQP--NYVMLACSRADHEP---------GDLLIVDNF-
1tig__c INVKEVRLS-PTIEEHDFNTKLRNARKFLEKG----DKVKAT--------------------------IRFKGRAITHKEIGQRVLDRLSEAC--ADIAVVE---------TAPKMDGRNMFLVLAPKND

