d1dzka_d1qd1b2
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1dzk_Ah
 |
d1dzka_ | A10-A25,A96-A157 | All beta proteins | Lipocalins | Lipocalins | Retinol binding protein-like | Odorant-binding protein |
1qd1_Bc
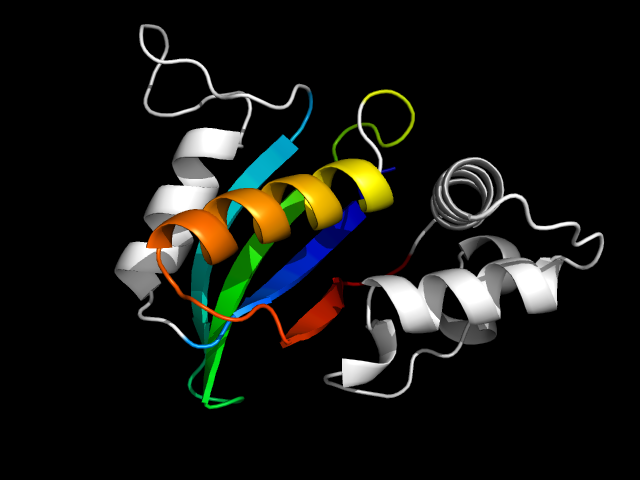 |
d1qd1b2 | B2181-B2210,B2211-B2326 | Alpha and beta proteins (a+b) | Ferredoxin-like | Formiminotransferase domain of formiminotransferase-cyclodeaminase. | Formiminotransferase domain of formiminotransferase-cyclodeaminase. | Formiminotransferase domain of formiminotransferase-cyclodeaminase.
|
Justification for analogy
The motif in 1dzk: the last strand and the helix come from C-termianl extension, and the remaining part comes from the lipocalin core.
Hybrid motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dzk_Ah felsgKWITsYIGSSD---------------------------TALIISNINVDeegDKTIMTGLLGKGTDI--EDQDLEKFKEVTRENGIPEENIVNIIErddcpa------------------------------------------------
1qd1_Bc -----FLLA-FNINLLstreqahrialdlreqgrgkdqpgrlkKVQAIGWYLDE---KNLAQVSTNLLDFEVtgLHTVFEETCREAQELSLPVVGSQLVGLvplkalldaaafycekenlfllqdehrirlvvnrlgldslapfkpkeriieylv
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dzk_Ah felsgkwITSYIGSSD---------------------------TALIISNINVDeegdKTIMTGLLGKgtdIEDQDLEKFKEVTRENG----iPEENIVNIIE----------------------------------------------rDDCP--a
1qd1_Bc ------fLLAFNINLLstreqahrialdlreqgrgkdqpgrlkKVQAIGWYLDE---kNLAQVSTNLL--dFEVTGLHTVFEETCREAqelslPVVGSQLVGLvplkalldaaafycekenlfllqdehrirlvvnrlgldslapfkpkeRIIEylv
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dzk_Ah ------------------------------------FELSG-------------KWI--TSYIGSSDTA---L-II--SN-IN--VD-------EEGDKTIMTGLLGKGTDIEDQDLEKFKEVTRENGIP-E----E--NI--V-NIIERDD-------------CPA
1qd1_Bc FLLAFNINLLSTREQAHRIALDLREQGRGKDQPGRLKKVQAIGWYLDEKNLAQVSTNLL--------DFEVTGLHTVFEETCREA-QELSLPVV---------GSQL-VGLVPLKALLDAAAFYCEKENLFLLQDEHRIRLVVNRLGLDSLAPFKPKERIIEYLV---
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dzk_Ah FELSGKWITS----YIGSSDT------------------------------ALIISNINVDEEGDK---TIMTGLLGKGTDIE------DQDLEKFKEVTRENGIPEEN--IVNIIERDDCPA-------------------------------------------------------
1qd1_Bc ----------FLLAFNIN---LLSTREQAHRIALDLREQGRGKDQPGRLKKVQAIGWYLD------EKNLAQVSTNLLD----FEVTGLHTVFEETCREAQELSLPV--VGSQLVG-------LVPLKALLDAAAFYCEKENLFLLQDEHRIRLVVNRLGLDSLAPFKPKERIIEYLV

