d1dzta_d1kpf__
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1dzt_Ah
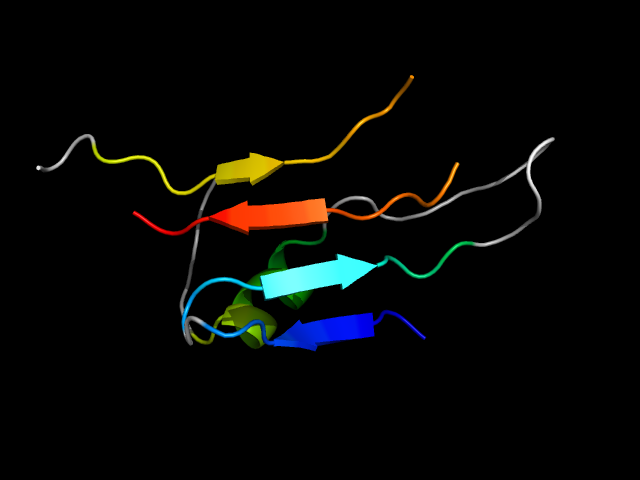 |
d1dzta_ | A1-A46,A67-A79,A107-A116 | All beta proteins | Double-stranded beta-helix | RmlC-like cupins | dTDP-sugar isomerase | dTDP-4-dehydrorhamnose 3,5-epimerase RmlC |
1kpf__c
 |
d1kpf__ | 16-89,90-106,107-126 | Alpha and beta proteins (a+b) | HIT-like | HIT-like | HIT (HINT, histidine triad) family of protein kinase-interacting proteins | Protein kinase C inhibitor-1, PKCI-1
|
Justification for analogy
The motif in 1dzt: comparing d1dzta_ and 2phla1 (different family same superfamily, since domain arrangement in 2phla1 is the same as subunit
arrangement in d1dzta_ and the curvature of the beta-sheets are very similar in these two proteins), the insertion is in different position relative to the core
structure. Hybrid motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dzt_Ah ---------------------MMIVIKTAip---DVLILEPKVfgde------rgfffesyNQQTFEELIGrkvtfgenAQGKLVRCAV-------ENKRQLWIPE--------
1kpf__c dtifgkiirkeipakiifeddRCLAFHDIspqapTHFLVIPKKhisqisvaedddesllghLMIVGKKCAAdlg---lnKGYRMVVNEGsdggqsvYHVHLHVLGGrqmhwppg
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dzt_Ah ---------------------MMIVIKTAIP---DVLILEPKV----fgDERGFF-fESYN-qQTFEELIGRkvtfgenAQGKLVRCAV-------ENKRQLWIPE--------
1kpf__c dtifgkiirkeipakiifeddRCLAFHDISPqapTHFLVIPKKhisqisVAEDDDesLLGHlmIVGKKCAAD---lglnKGYRMVVNEGsdggqsvYHVHLHVLGGrqmhwppg
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dzt_Ah ---------------------MMIVIKTA-I--PDVLILEPKVFGD---ERG-FF--FES--YNQQTFEELI-G-RKVTFGENAQGKLVRCAV-------ENKRQLWIP-E-------
1kpf__c DTIFGKIIRKEIPAKIIFEDDRCLAFHDISPQAPTHFLVIPKKHISQISVAEDDDESLLGHLMI--VGKKCAADLGLN-----KGYRMVVNEGSDGGQSVYHVHLHVLGGRQMHWPPG
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1dzt_Ah ---------------------MMIVIKTAIP---DVLILEPKVFGDERGFFFESYN--------------------QQTFEELIGRKVTFGENA------QGKLVRCAV--------ENKRQLWIPE--------
1kpf__c DTIFGKIIRKEIPAKIIFEDDRCLAFHDISPQAPTHFLVIPK--------------KHISQISVAEDDDESLLGHLMIVGKKCAA---------DLGLNKGYRMVVNE-GSDGGQSVYHVHLHVLGGRQMHWPPG

