d1e3pa3d1gpqa_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1e3p_Ah
 |
d1e3pa3 | A3-A57,A116-A151 | Alpha and beta proteins (a+b) | Ribosomal protein S5 domain 2-like | Ribosomal protein S5 domain 2-like | Ribonuclease PH domain 1-like | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4 |
1gpq_Ac
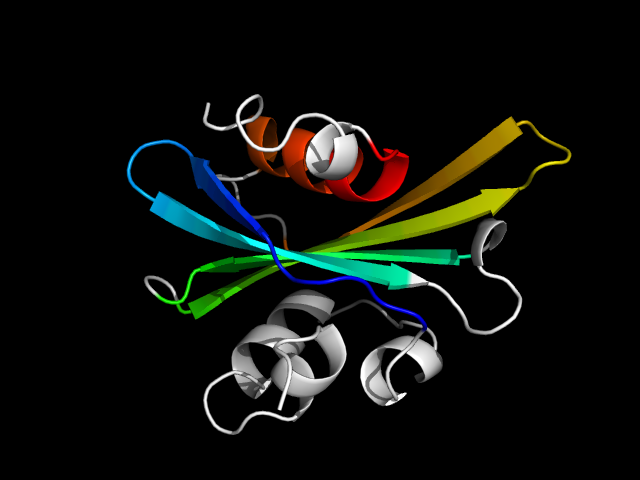 |
d1gpqa_ | A2-A89,A90-A128 | Alpha and beta proteins (a+b) | Inhibitor of vertebrate lysozyme, Ivy | Inhibitor of vertebrate lysozyme, Ivy | Inhibitor of vertebrate lysozyme, Ivy | Inhibitor of vertebrate lysozyme, Ivy
|
Justification for analogy
The motif in 1e3p: the first two strands come from insertion, and the remaining part comes from the core. Hybrid motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1e3p_Ah -----------------------------------NETHYAEAVIDngafgTRTIRFETGrla--rqaAGSAVAYLD-dDTMVLSATTASKNPNEIQVVATImalnpdhlYDVVAINAASASTQlaglp------
1gpq_Ac ddltisslakgettkaafnqmvqghklpawvmkggTYTPAQTVTLG-----DETYQVMSAckphdcgsQRIAVMWSEksNQMTGLFSTIDEKTSQEKLTWLNv---ndalSIDGKTVLFAALTGslenhpdgfnf
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1e3p_Ah --------------------------------NETHYA-EAVIDNGaFGTRTIRFETGRlarqaAGSAVAYLD-----dDTMVLSATTASknpnEIQVVATIMALNPDH---LYDVVAINAAS--astqlaglp
1gpq_Ac ddltisslakgettkaafnqmvqghklpawvmKGGTYTpAQTVTLG-DETYQVMSACKP--hdcGSQRIAVMWseksnqMTGLFSTIDEK----TSQEKLTWLNVNDALsidGKTVLFAALTGslenhpdgfnf
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1e3p_Ah -----------------------------N---ETHYA-EAVIDNGAFGTRTIRFETGRLARQAAGSAVAYLD-----DDTMVLSATTASKNPNEIQVVATIMALNPDHL--YDVVAINAASA-----------STQLAGLP
1gpq_Ac DDLTISSLAKGETTKAAFNQMVQGHKLPAWVMKGGTYTPAQTVTL-GDETYQVMSACKP--HDCGSQRIAVMWSEKSNQMTGLFSTIDEK----TSQEKLTWLNVNDALSIDGKTVLFAALTGSLENHPDGFNF--------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1e3p_Ah -------------------------------------NETHYAEAVIDNGAFGT-RTIRFETGRLARQAA----------GSAVAYL-DDDTMVLSATTASKNPNEIQVVATIMALNPDHLY---DVVAINAASASTQLAGLP-------------
1gpq_Ac DDLTISSLAKGETTKAAFNQMVQGHKLPAWVMKGGTYTPAQTVTLG--------DETYQVMS--------ACKPHDCGSQRIAVMWSEKSNQMTGLFSTIDEKTSQEKLTWL----------NVNDALSIDGKTVLFAAL---TGSLENHPDGFNF

