d1efdn_d1r0va3
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1efd_Nh
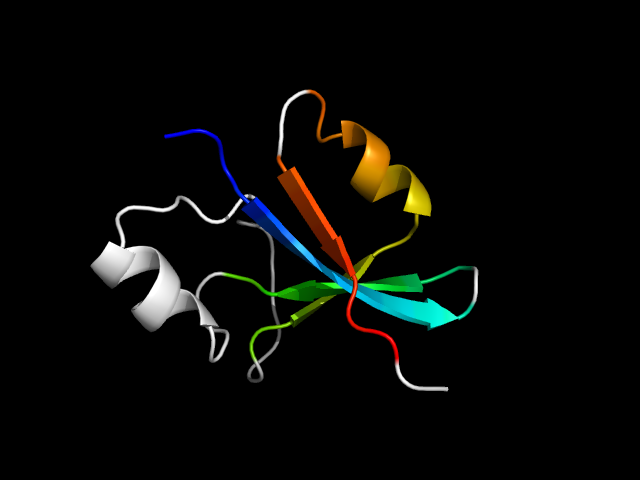 |
d1efdn_ | N174-N241 | Alpha and beta proteins (a/b) | Chelatase-like | "Helical backbone" metal receptor | Periplasmic ferric siderophore binding protein FhuD | Periplasmic ferric siderophore binding protein FhuD |
1r0v_Ac
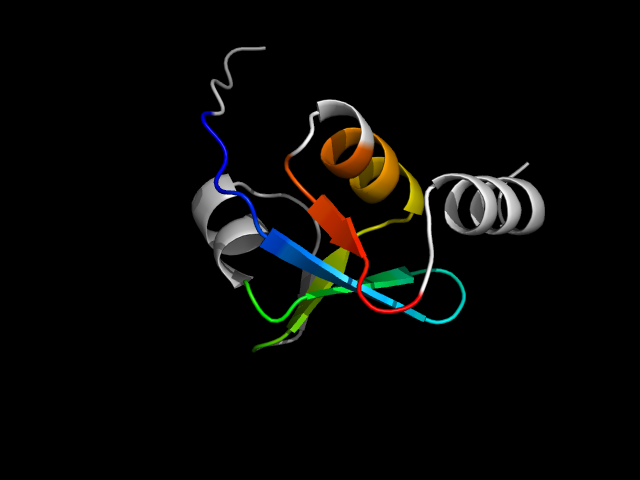 |
d1r0va3 | A140-A214 | Alpha and beta proteins (a+b) | MutS N-terminal domain-like | tRNA-intron endonuclease N-terminal domain-like | tRNA-intron endonuclease N-terminal domain-like | Dimeric tRNA splicing endonuclease, domains 1 and 3
|
Justification for analogy
Comparing the two duplicates in 1efd, the motif results from some deletions in the Rossmann-like structure. Strictly speaking, this is not a hybrid motif.
However, since d1r0va3 (and its homolog d1w7aa4) is drastically different than Rossmann structures, the motif in 1efd and the motif in 1r0v are analogs.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1efd_Nh -----ARPLLLTTLIDpRHMLVFGpnslfqeildeygipnawqgetnfwGSTAVSIDRLAAYKd-vDVLCFDhd-------------
1r0v_Ac geqkeELPEIAGVLSD-EYVITKQteifsry-----------fygsekgDLVTLSLIESLYLLdlgKLNLLNadreelvkrarever
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1efd_Nh -----ARPLLLTTLIDpRHMLVFGpnslfqeildeygipnaWQGE-------TNFWgSTAVSIDRLAAY-KDVDVLCFD-------------hd
1r0v_Ac geqkeELPEIAGVLSD-EYVITKQ-----------------TEIFsryfygsEKGD-LVTLSLIESLYLlDLGKLNLLNadreelvkrarever
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1efd_Nh -----ARPLLLTTLIDPRHMLVF-GPNSLFQEILDEYGIPNAWQ---G--ETNFWGSTAVSIDRL-AAY-KDVDVLCFD---------------HD
1r0v_Ac GEQKEELPEIAGVLSD-EYVITKQTE----------------IFSRYFYGSEKG-DLVTLSLIESLY-LLDLGKLNLLNADREELVKRAREVER--
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1efd_Nh ARP--------LLLTTLIDPRHMLVFGPNSLFQEILDEYGIPNAWQ----------GETNFWGSTAVSIDRLAA-YKDVDVLCFDHD----------------
1r0v_Ac ---GEQKEELPEIAGVLSD-EYVITK--------------------QTEIFSRYFYGSEKG-DLVTLSLIESLYLLDLGKLNLL---NADREELVKRAREVER

