d1eg3a1d1paqa_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1eg3_Ai
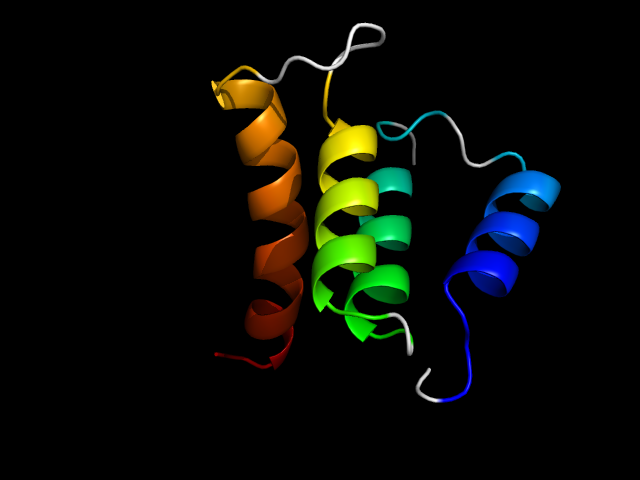 |
d1eg3a1 | A121-A144,A195-A250 | All alpha proteins | EF Hand-like | EF-hand | EF-hand modules in multidomain proteins | Dystrophin |
1paq_Ac
 |
d1paqa_ | A544-A582,A583-A704 | All alpha proteins | alpha-alpha superhelix | ARM repeat | MIF4G domain-like | Translation initiation factor eIF-2b epsilon
|
Justification for analogy
In 1eg3, helix 1 and 4 in the first EF domain and helix 1 and 2 in the second EF domain form a right-handed superhelix, which is an interface motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1eg3_Ai --------------ldLLSLSAACDALDQHNl---kQNDqpRVLSFKTGIISLCk----------------aHLEDKYRYLFKQVAsstgfcDQRRLGLLLHDSIQIPRQLGE--------------------------------------------------------
1paq_Ac dfekegiatveramenNHDLDTALLELNTLRmsmnvTYH--EVRIATITALLRRvyhfiatqtlgpkdavvkVFNQWGLLFKRQAF------DEEEYIDLMNIIMEKIVEQSFdkpdlilfsalvslydndiieedviykwwdnvstdprydevkkltvkwvewlqnad
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1eg3_Ai -------------lDLLS-LSAACDALDQHN---lKQNDqpRVLSFKTGIISL----------------ckaHLEDKYRYLFKQVAsstgfcDQRRLGLLLHDSIQ-IPRQLGE-------------------------------------------------------
1paq_Ac dfekegiatveramENNHdLDTALLELNTLRmsmnVTYH--EVRIATITALLRrvyhfiatqtlgpkdavvkVFNQWGLLFKRQAF------DEEEYIDLMNIIMEkIVEQSFDkpdlilfsalvslydndiieedviykwwdnvstdprydevkkltvkwvewlqnad
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1eg3_Ai LDLLSLSAACDALDQHNLKQNDQ-PRVLSFKTGIISLCKAHLEDKYRYLFKQVASSTGFCDQRRLGLLLHDSIQIPRQLG------E-------------------------------------------------------------------------------------------------
1paq_Ac ---------------------DFE-KEGIATVERAMENNHDLDTALLELNTL-RMSMNVTYHEVRIATITALLRRVYHFIATQTLGPKDAVVKVFNQWGLLFKRQAFDEEEYIDLMNIIMEKIVEQSFDKPDLILFSALVSLYDNDIIEEDVIYKWWDNVSTDPRYDEVKKLTVKWVEWLQNAD
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1eg3_Ai LDLLS--LSAACDALDQHNLKQNDQPR---------------------------------------------------------VLSFKTGIISLCKA---HLEDKYRYLFKQVASSTGFCD-----------QRRLGLLLHDSIQIPRQLGE---------------------------------------
1paq_Ac -----DFEKEGIATVERA---------MENNHDLDTALLELNTLRMSMNVTYHEVRIATITALLRRVYHFIATQTLGPKDAVVKVFNQWGLLFKR---QAFDEEEYIDLMNIIM--------EKIVEQSFDKPDLILFSALVSLYDN------DIIEEDVIYKWWDNVSTDPRYDEVKKLTVKWVEWLQNAD

