d1ert__d1jr3d2
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1ert__h
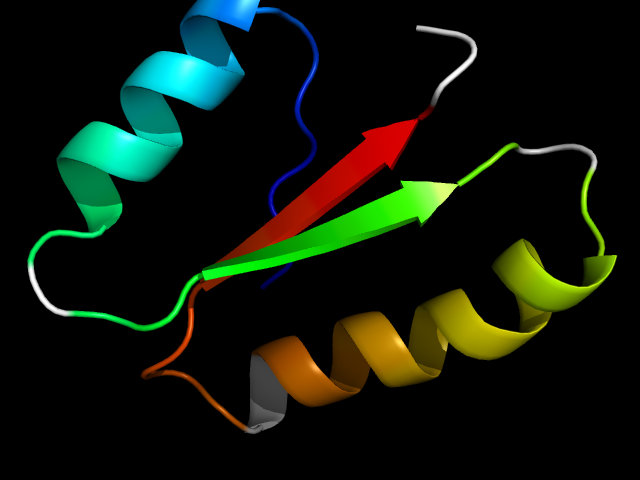 |
d1ert__ | 1-61 | Alpha and beta proteins (a/b) | Thioredoxin fold | Thioredoxin-like | Thioltransferase | Thioredoxin |
1jr3_Dc
 |
d1jr3d2 | D17-D117 | Alpha and beta proteins (a/b) | P-loop containing nucleoside triphosphate hydrolases | P-loop containing nucleoside triphosphate hydrolases | Extended AAA-ATPase domain | delta subunit of DNA polymerase III, N-domain
|
Justification for analogy
The motif in 1ert: comparing d1ert__ and d1h75a_ (same family, different protein), one strand and one helix are insertion. hybrid.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ert__h --MVKQIE------SKTAFQEALDAAG--------------------------------dKLVVVDFSAt--wCGPCKMIKPFFHSLSekYSNVIFLEVDvdd
1jr3_Dc lrAAYLLLgndpllLQESQDAVRQVAAaqgfeehhtfsidpntdwnaifslcqamslfasRQTLLLLLPengpNAAINEQLLTLTGLL--HDDLLLIVRGnkl
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ert__h --MVKQI------eSKTAFQEALDAAG--------------------------------dKLVVVDFSA--TWCGPCKMIKPFFHSLSekYSNVIFLEVDVDD
1jr3_Dc lrAAYLLlgndpllLQESQDAVRQVAAaqgfeehhtfsidpntdwnaifslcqamslfasRQTLLLLLPenGPNAAINEQLLTLTGLL--HDDLLLIVRGNKL
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ert__h --MVKQIE------SKTAFQEALDAAG--DK------------------------------LVVVDFSA--TWCGPCKMIKPFFHSLSEKYSNVIFLEVDVDD--
1jr3_Dc LRAAYLLLGNDPLLLQESQDAVRQVAAAQGFEEHHTFSIDPNTDWNAIFSLCQAMSLFASRQTLLLLLPENGPNAAINEQLLTLTGLL--HDDLLLIVRG--NKL
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ert__h M---VKQIES------KTAFQEALDAAGDK------LVVVDFSATWCGPCKMIKPFFHSLSEKYSN-VIFLEVDVDD--------------------------------
1jr3_Dc -LRAAYLLLGNDPLLLQESQDAVRQVA---AAQGFEEHHTFSIDPNTD--WNAIFSLCQAMSLFASRQTLLLLLP--ENGPNAAINEQLLTLTGLLHDDLLLIVRGNKL

