d1gx3a_d1epaa_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1gx3_Ah
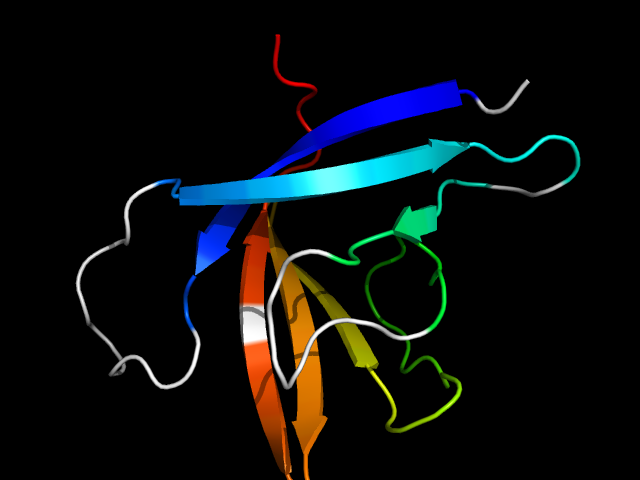 |
d1gx3a_ | A85-A185 | Alpha and beta proteins (a+b) | Cysteine proteinases | Cysteine proteinases | Arylamine N-acetyltransferase | Arylamine N-acetyltransferase |
1epa_Ac
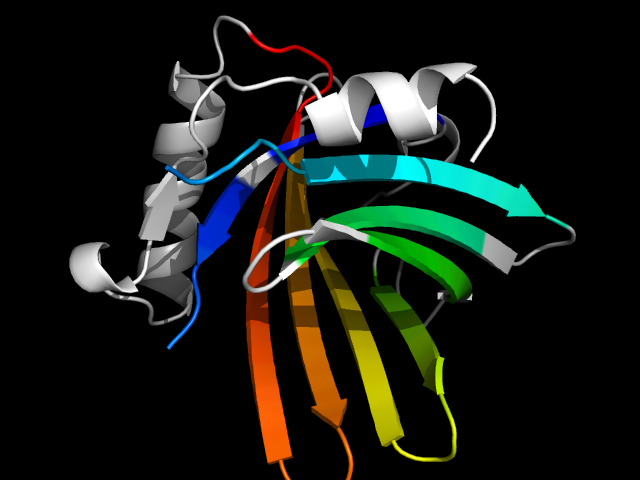 |
d1epaa_ | A3-A162 | All beta proteins | Lipocalins | Lipocalins | Retinol binding protein-like | Retinoic acid-binding protein
|
Justification for analogy
The motif in 1gx3: compare d1gx3a_ with classic cysteine proteinase papain d1ppn__ (same superfamily), half of the barrel in 1gx3 is
absent in 1ppn. So this is a hybrid motif and is analog to true barrels in all-beta class. Note that the cysteine proteinase superfamily is
very diverged in structures.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1gx3_Ah --------gfEVERL-SGRVVwmraddaplpaQTHNVLSVAVPGa--dGRYLVDVgfggqtltSPIRLEA-------GPVQQTRHEPYRLTRHG------DDHTLAAQVRGEWQPlYTFTTEPRP-----------------------------------------
1epa_Ac vkdfdiskflGFWYEiAFASKmgtp--glahkEEKMGAMVVELKenllALTTTYYse---dhcVLEKVTAtegdgpaKFQVTRLSGKKEVVVEAtdyltyAIIDITSLVAGAVHR-TMKLYSRSLddngealynfrkitsdhgfsetdlyilkhdltcvkvlqsaa
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1gx3_Ah ------gfeVERLSG-RVVWMraddaplpaq--THNVLSVAVpgADGRYLVDvGFGG--qtltSPIR-----leAGPVQQTRHEPYRLTRHG------DDHTLAAQVRGEWQPLYTFTTEPRP----------------------------------------
1epa_Ac vkdfdiskfLGFWYEiAFASKmgtpglahkeekMGAMVVELK--ENLLALTT-TYYSedhcvlEKVTategdgpAKFQVTRLSGKKEVVVEAtdyltyAIIDITSLVAGAVHRTMKLYSRSLDdngealynfrkitsdhgfsetdlyilkhdltcvkvlqsaa
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1gx3_Ah ---------GFEVERLSGRVVWMR----------ADDAPLPAQTHNVLSVAVPGADGRYLVDVGFGG---QTLTSPIRLE----AGPVQQTRHEPYRLTRHGD------DHTLAAQVRGEWQPLYTFTTEPRP-----------------------------------------
1epa_Ac VKDFDISKF--LGFWYEIAFASKMGTPGLAHKEE---------KMGAMVVEL-KENLLALTTTYYSEDHC-VLEKVTATEGDGPAKFQVTRLSGKKEVVVEATDYLTYAIIDITSLVAGAVHRTMKLY-SRSLDDNGEALYNFRKITSDHGFSETDLYILKHDLTCVKVLQSAA
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1gx3_Ah GFEV----------ERLSG-RVVWMRADDAPLPAQT--------------HNVLSVAVPGADGRYLVDVGFGGQTLT---------SPIRLEA-----GPVQQTRHEPYRLTRHGD-------DHTLAAQVRGEWQPL-YTFTTEPRP-------------------------------------------
1epa_Ac ----VKDFDISKFLGFWYEIAFAS------------KMGTPGLAHKEEKMGAMVVELKE--NLLALTTT--------YYSEDHCVLEKVTATEGDGPAKFQVTRLSGKKEVVVEA-TDYLTYAIIDITSLVAGAVHRTMKLYSR----SLDDNGEALYNFRKITSDHGFSETDLYILKHDLTCVKVLQSAA

