d1gxja_d1p5dx3
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1gxj_Ai
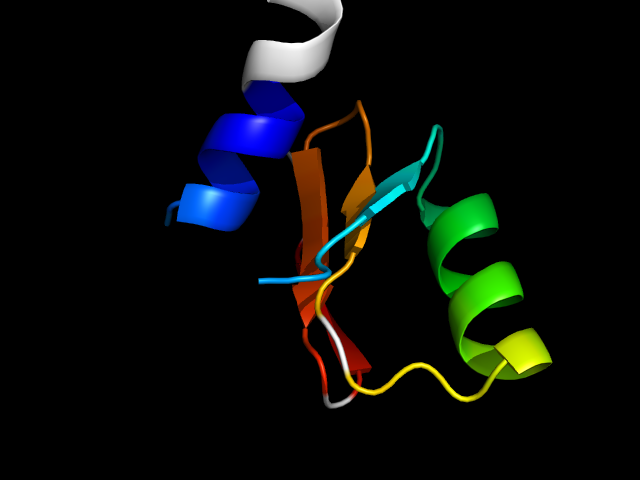 |
d1gxja_ | A527-A539,A615-A658 | Alpha and beta proteins (a+b) | Smc hinge domain | Smc hinge domain | Smc hinge domain | Smc hinge domain |
1p5d_Xc
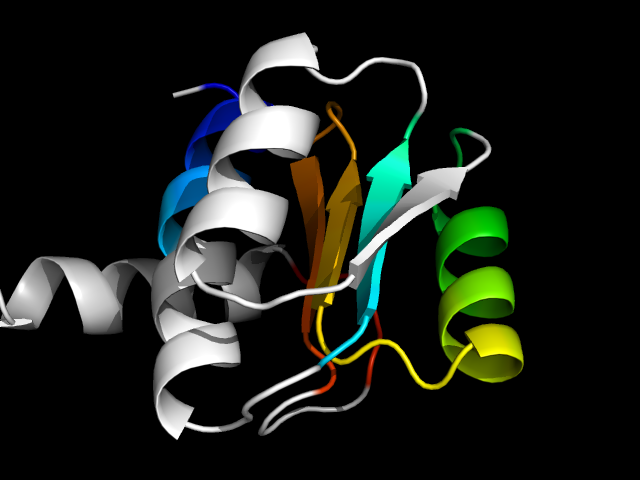 |
d1p5dx3 | X258-X276,X277-X356 | Alpha and beta proteins (a/b) | Phosphoglucomutase, first 3 domains | Phosphoglucomutase, first 3 domains | Phosphoglucomutase, first 3 domains | Phosphomannomutase/phosphoglucomutase
|
Justification for analogy
1gxj (Smc hinge domain) appears to have two duplicates. Each duplicate has three strands linked by two helices. The two duplicates swap the
helix between strand 1 and strand 2. So, although the SCOP fold "Smc hinge domain" only has one protein, I think its core is different from 1p5d,
which is apparently a mini-Rossmann. Thus these two motifs are analogs.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1gxj_Ai dekysLAVSVLLG-----------GNSVVV----------------------ETLDDAIRMKKKYRLNtRIATLDGELISG---rGAITggre-----------
1p5d_Xc ----iYPDRLLMLfakdvvsrnpgADIIFDvkctrrlialisgyggrpvmwkTGHSLIKKKMKETGAL-LAGEMSGHVFFKerwfGFDDgiysaarlleilsqd
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1gxj_Ai DEKYSLAVSVLLG-------GNSVVV----------------------eTLDDAIRMKKKYRLnTRIATLDGELISG---RGAITGG-----------re
1p5d_Xc IYPDRLLMLFAKDvvsrnpgADIIFDvkctrrlialisgyggrpvmwktGHSLIKKKMKETGA-LLAGEMSGHVFFKerwFGFDDGIysaarlleilsqd
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1gxj_Ai -DEKYSLAVSVL-LG----G------NSVVV----------------------ETLDDAIRMKKKYRLNTRIATLDGELISG--R-GAITGG-------------RE
1p5d_Xc I----YPDRL-LMLFAKDVVSRNPGADIIFDVKCTRRLIALISGYGGRPVMWKTGHSLIKKKMKETGA-LLAGEMSGHVFFKERWFGFDDGIYSAARLLEILSQD--
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1gxj_Ai DEK---YSLAVSVLLGG--------NSVVV----------------------ETLDDAIRMKKKYRLNTRIATLDGELISG----RGAITGGRE--------------
1p5d_Xc ---IYPDRLLMLFAKD-VVSRNPGADIIFDVKCTRRLIALISGYGGRPVMWKTGHSLIKKKMKETGAL-LAGEMSGHVFF-KERWFGFDDG---IYSAARLLEILSQD

