d1h0hb_d1hz6b_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1h0h_Bh
 |
d1h0hb_ | B1-B14,B157-B214 | Alpha and beta proteins (a+b) | Ferredoxin-like | 4Fe-4S ferredoxins | Ferredoxin domains from multidomain proteins | Tungsten containing formate dehydrogenase, small subunit |
1hz6_Bc
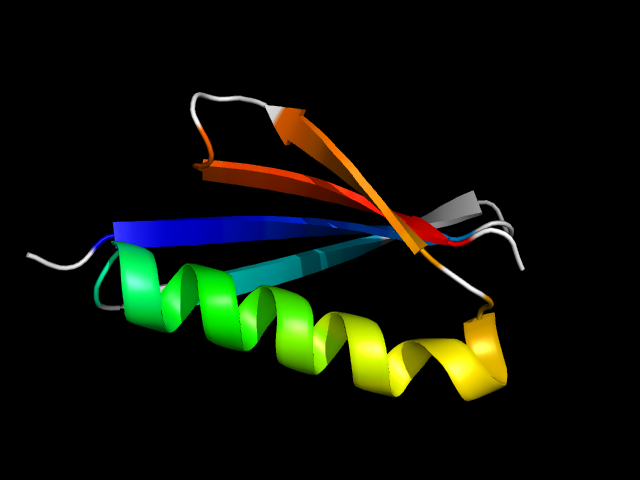 |
d1hz6b_ | B2-B13,B14-B64 | Alpha and beta proteins (a+b) | beta-Grasp (ubiquitin-like) | Immunoglobulin-binding domains | Immunoglobulin-binding domains | Immunoglobulin light chain-binding domain of protein L
|
Justification for analogy
The motif in 1h0h: hybrid, see the "structural drift" paper.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1h0h_Bh --SKGFFVDTTrctac-ptgAMNFG--DLSEMEAMASARLAEIKAAYSdAKLCDpddVRVIFLTAhnpklyheyava
1hz6_Bc eeVTIKANLIFa----ngstQTAEFkgTFEKATSEAYAYADTLKKDNGeWTVDVadkGYTLNIKFag----------
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1h0h_Bh -SKGFFVDTTrctacptgAMNFGD-LSEMEAMASARLAEIKAAYSDAKLCDpdDVRVIFLTAHnpklyheyava
1hz6_Bc eEVTIKANLI-fangstqTAEFKGtFEKATSEAYAYADTLKKDNGEWTVDVadKGYTLNIKFA----------g
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1h0h_Bh --SKGFFVDTT---RCTACPTGAMNFGD--L-SEME-AMASA-RLAE-IKAAYSDAKLCDP-DDVRVIFLTAH-NPKLYHEYAVA
1hz6_Bc EEVTIKANLIFANGST------QTAEFKGTFEKA-TSEA-YAYA-DTLKKDNGE-WTVDVAD-KGYTLNIKFAG-----------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1h0h_Bh SKGFFVDTTRCTACPTGA-----------MNFGDLSEMEAMASARLAEIKAAYSDAKLCDPDD----VRVIFLTAHNPKLYHEYAVA--
1hz6_Bc EEVTIKAN----------LIFANGSTQTAEFKGTFEKATSEAYAYADTLKKDNGEWTVD----VADKGYTLNIKF------------AG

