d1h6da2d1kjka_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1h6d_Ah
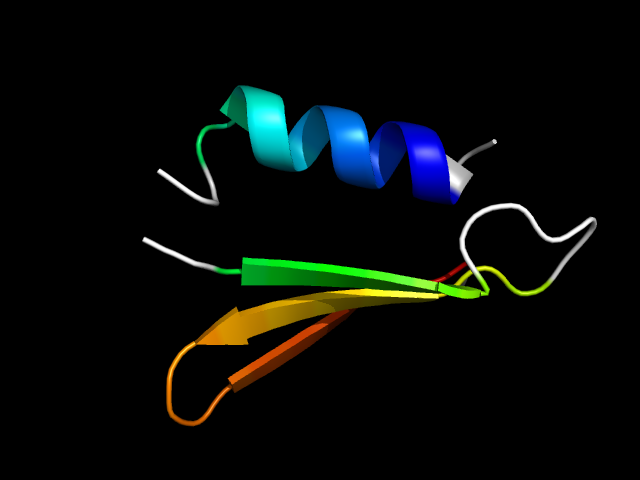 |
d1h6da2 | A213-A229,A337-A367 | Alpha and beta proteins (a+b) | Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain | Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain | Glucose 6-phosphate dehydrogenase-like | Glucose-fructose oxidoreductase |
1kjk_Ac
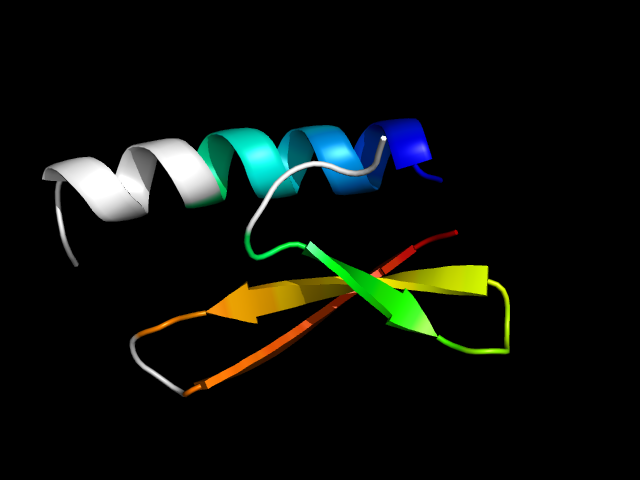 |
d1kjka_ | A40-A59,A11-A39 | Alpha and beta proteins (a+b) | DNA-binding domain | DNA-binding domain | lambda integrase N-terminal domain | lambda integrase N-terminal domain
|
Justification for analogy
The motif in 1h6d: compared to d1lc0a2 (same superfamily), two strands are insertion. Hybrid. See d1drw_2 for the consensus of this fold as described
in SCOP.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1h6d_Ah dpMNRAAVKLIRENqlg-------kaVLLMDPatgyyQNLISVQTP--GHANQSMMp
1kjk_Ac --DRRIAITEAIQAnielfsghdlppNLYIRN-----NGYYCYRDPrtGKEFGLGR-
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1h6d_Ah --DPMNRAAVKLIReNQLGK--avlLMDPatGYYQNLISVQTpgHANQSMM-p
1kjk_Ac drRIAITEAIQANI-ELFSGhdlppNLYI-rNNGYYCYRDPR--TGKEFGLgr
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1h6d_Ah --DPMNRAAVKLIRENQLG-KA---VLLMDP---ATGYYQNLISVQTPGH-ANQSMM-P
1kjk_Ac DRRIAITEAIQANI--ELFSGHDLP-PNLYIRNN----GYYCYRDPR--TGKEFGLGR-
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1h6d_Ah DPMNRAAVKLIRENQLGKAV----------LLMDPATGYYQ-NLISVQTP--GHANQSMMP-
1kjk_Ac --DRRIAITEAIQ-ANIE--LFSGHDLPPNLYIRN------NGYYCYRDPRTGKEFGLG--R

