d1hrda1d1rp3a3
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1hrd_Ah
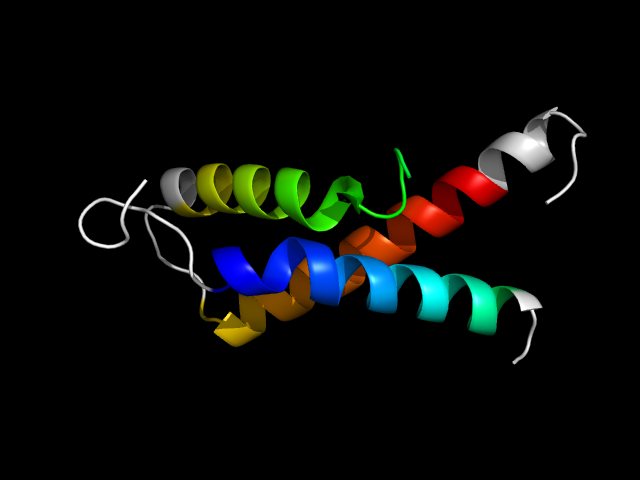 |
d1hrda1 | A195-A226,A369-A424 | Alpha and beta proteins (a/b) | NAD(P)-binding Rossmann-fold domains | NAD(P)-binding Rossmann-fold domains | Aminoacid dehydrogenase-like, C-terminal domain | Glutamate dehydrogenase |
1rp3_Ac
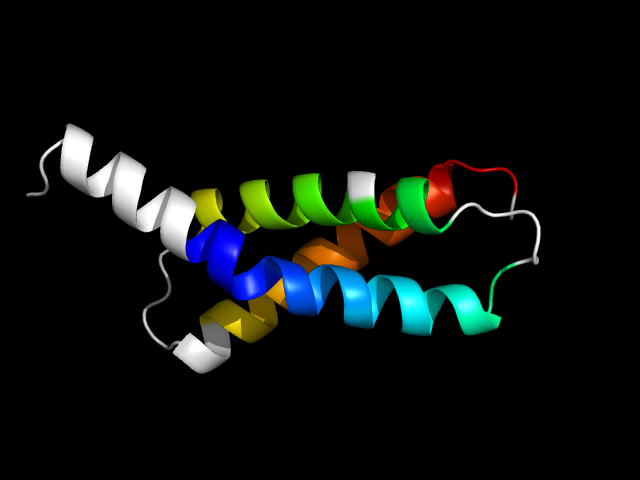 |
d1rp3a3 | A1-A36,A37-A87 | All alpha proteins | Sigma2 domain of RNA polymerase sigma factors | Sigma2 domain of RNA polymerase sigma factors | Sigma2 domain of RNA polymerase sigma factors | Sigma factor sigma-28 (FliA)
|
Justification for analogy
The motif in 1hrd: first helix is from the Rossmann core, the other two helices are from C-terminal extension. So this is a hybrid motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1hrd_Ah ----karsfggslVRPEATGYGSVYYVEAVMKHend--SKAVN-AGGVLVSGFEMSQnserlswTAEEVDSKLHQVMTDIHDGSAaaaeryglgy
1rp3_Ac mknpysnqiereeLILKYLPLVKAIATNIKKHLpedvdIRDLIsYGVIGLIKAVDNLstenpkrAEAYIKLRIKGAIYDYLRSLDfg--------
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1hrd_Ah kARSF-ggslvRPEATGYGSVYYVEAVMKHEND----------sKAVNA-GGVLVSGFEmsqnserlsWTAEEVDSKLHQVMTDIHDGSAAAAERYglgy
1rp3_Ac -MKNPysnqieREELILKYLPLVKAIATNIKKHlpedvdirdliSYGVIgLIKAVDNLS--------tENPKRAEAYIKLRIKGAIYDYLRSLDFG----
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1hrd_Ah --------KAR-SFGGSLVRPEATGYGSVYYVEAVM-----KHENDSKAVNA-GGVLVSGFEMSQNSERLS----WTAEEVDSKLHQVMTDIHDGSAAAAERYGLGY
1rp3_Ac MKNPYSNQIERE--ELILKYLPLVKAIATNIKKHLPEDVDI-----R-DLISYGVIGLIKAVDNL----STENPKRAEAYIKLRIKGAIYDYLRSLDF--G------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1hrd_Ah KARSFGGSLV-------RPEATGYGSVYYVEAVMKHENDSKA-------------------VNAGGVLVSGFEMSQNSERLSW---TAEEVDSKLHQVMTDIHDGSAAAAERYGLGY---
1rp3_Ac ----------MKNPYSNQIEREELILKYLPLVKAIAT-----NIKKHLPEDVDIRDLISYGVIGLIKAVDNL-----------STENPKRAEAYIKLRIKGAIYDYLRSL-------DFG

