d1i9ea_d1mzka_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1i9e_Ah
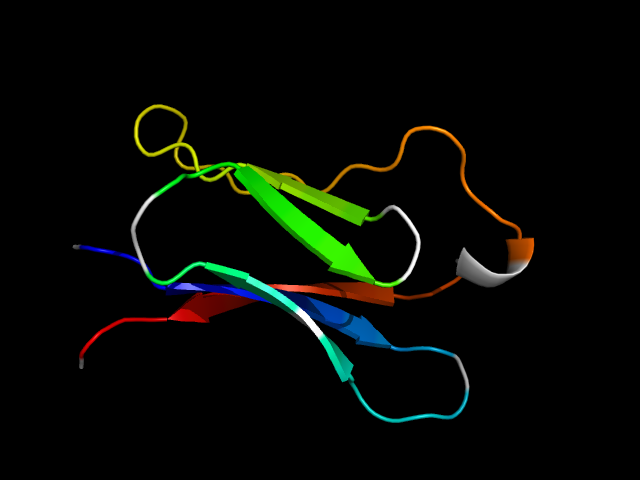 |
d1i9ea_ | A28-A96 | All beta proteins | Immunoglobulin-like beta-sandwich | Immunoglobulin | V set domains (antibody variable domain-like) | T-cell antigen receptor |
1mzk_Ah
 |
d1mzka_ | A177-A298 | All beta proteins | SMAD/FHA domain | SMAD/FHA domain | FHA domain | Kinase associated protein phosphatase
|
Justification for analogy
The motif in 1i9e: two strands don't belong to the Ig core; hybrid motif.
Although 1mzk is also Ig-like (the first strand in Ig fold is the 5th strand in 1mzk, according to SCOP's 11 strands numbering), the motif
in 1i9e and the motif in 1mzk can not be homologs: if strands in Ig fold is counted from 1 to 7, all other strands are labeled as i
(insertion), the motif in 1i9e is 34ii56 while the motif in 1mzk is iii127. So no homologous elements are superimposed.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1i9e_Ah -aTPYLFWYVQYpRQGLQLlLKYYs---gDPVVQGVn----------------gFEAEFSKSNSSFHLRKASVHW---------------------------------sdSAVYFCAVSGFa-
1mzk_Ah lgSSWLFLEVIAgPAIGLQ-HAVNstsssKLPVKLGrvspsdlalkdsevsgkhAQITWNSTKFKWELVDMGSLNgtlvnshsishpdlgsrkwgnpvelasddiitlgtTTKVYVRISSQne
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1i9e_Ah -ATPYLFWYVQYPRQGLQLlLKYY---SGDPVVQGV----------------NGFEAEFSKSNSSFHLRKASVHW---------------------------------SDSAVYFCAVSGFA-
1mzk_Ah lGSSWLFLEVIAGPAIGLQ-HAVNstsSSKLPVKLGrvspsdlalkdsevsgKHAQITWNSTKFKWELVDMGSLNgtlvnshsishpdlgsrkwgnpvelasddiitlGTTTKVYVRISSQNe
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1i9e_Ah -ATPYLFWYVQYP-RQGLQLLLKYY--SGDPVVQGVN----------------G-FEAEFSKSNSSFHLRKA-S--V-------------------------------HWSDSAVYFCAVSGFA
1mzk_Ah LGSSWLFLEVIAGP-AIGLQ-HAVNSTSSSKLPVKLGRVSPSDLALKDSEVSGKHAQITWNSTKFKWELVDMGSLNGTLVNSHSISHPDLGSRKWGNPVELASDDIITLGTTTKVYVRISSQNE
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1i9e_Ah A--TPYLFWYVQYP--RQGLQLLLKYYSG---DPVVQGVN------------------GFEAEFSKSNSSFHLRKASVHWSDS-------------------------------------AVYFCAVSGFA--
1mzk_Ah -LGSSWLFLEVI--AGPAIGLQ-HAVNSTSSSKLPVKL--GRVSPSDLALKDSEVSGKHAQITWNSTKFKWELVDMGSL----NGTLVNSHSISHPDLGSRKWGNPVELASDDIITLGTTTKVYVRISSQ-NE

