d1izna_d1gkza1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1izn_Ai
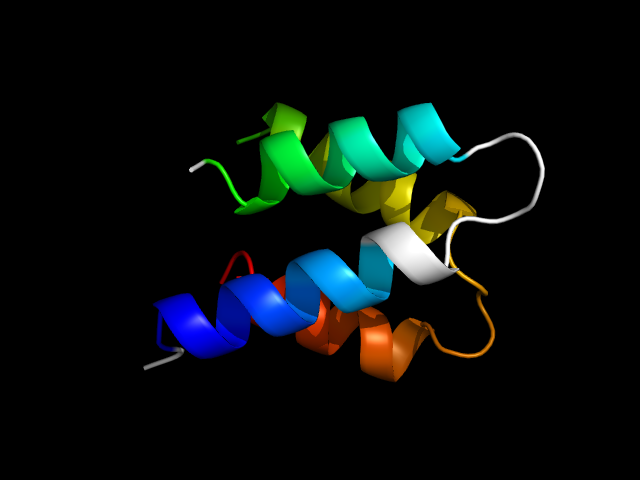 |
d1izna_ | A7-A42,B2-B33 | Multi-domain proteins (alpha and beta) | Subunits of heterodimeric actin filament capping protein Capz | Subunits of heterodimeric actin filament capping protein Capz | Capz alpha-1 subunit | Capz alpha-1 subunit |
1gkz_Ac
 |
d1gkza1 | A64-A104,A127-A174 | All alpha proteins | Bromodomain-like | alpha-ketoacid dehydrogenase kinase, N-terminal domain | alpha-ketoacid dehydrogenase kinase, N-terminal domain | Branched-chain alpha-ketoacid dehydrogenase kinase (BCK)
|
Justification for analogy
In 1izn, the 4-helix bundle uses corresponding parts in the two chains. interface motif. Note that the two chains are not identical, but they are in the same superfamily in SCOP.
In 1gkz, the 4-helix bundle is the core motif of that domain.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1izn_Ai -----rvSDEEKVRIAAKFithappgeFNEVFNDVRLLLNn-------SDQQLDCALDLMRRL-----PPQQIEKNLSDLIDLVP------
1gkz_Ac lqqelpvRIAHRIKGFRSLpfii--gcNPTILHVHELYIRafqqllddHKDVVTLLAEGLRESrkhieDEKLVRYFLDKTLTSRLgirmla
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1izn_Ai --rvSDEEKVRIAAKFITHAPP-gEFNEVFNDVRLLLNN-------SDQQLDCALDLMR-----rlpPQQIEKNLSDLIDLVP------
1gkz_Ac lqqeLPVRIAHRIKGFRSLPFIigCNPTILHVHELYIRAfqqllddHKDVVTLLAEGLResrkhiedEKLVRYFLDKTLTSRLgirmla
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1izn_Ai R--VS-DEEKVRIAAKFITHAPPGEFNE-VFNDVRLLL-NN--------SDQQLDCALDLMR-R---LP-PQQ-IEKNLSDLI-DLVP------
1gkz_Ac LQQ-ELPVRIAHRIKGFRSLPFIIGCNPTILHVHEL-YIRAFQQLLDDHK-DVVTLLAEGLRESRKHIEDEKLVR-YFLDKTLTS-RLGIRMLA
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1izn_Ai RVSDEEKVRIAAKFITH---APPGE-----FNEVFNDVRLLLNN---SDQQLDCALDLMRRLPPQ------------QIEKNLSDLIDLVP-------
1gkz_Ac --LQQELPVRIAHRIKGFRSLPFIIGCNPTILHVHELYIRAF--QQLLDDHKDVVTLLAEG----LRESRKHIEDEKLVRYFLDKTLTSR-LGIRMLA

