d1j22a_d1m2aa_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1j22_Ah
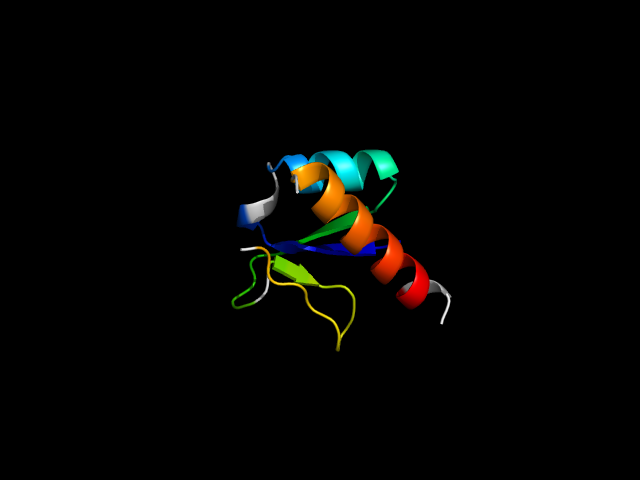 |
d1j22a_ | A5-A50,A116-A135 | Alpha and beta proteins (a/b) | Restriction endonuclease-like | Restriction endonuclease-like | XPF/Rad1/Mus81 nuclease | Putative ATP-dependent RNA helicase Hef, nuclease domain |
1m2a_Ac
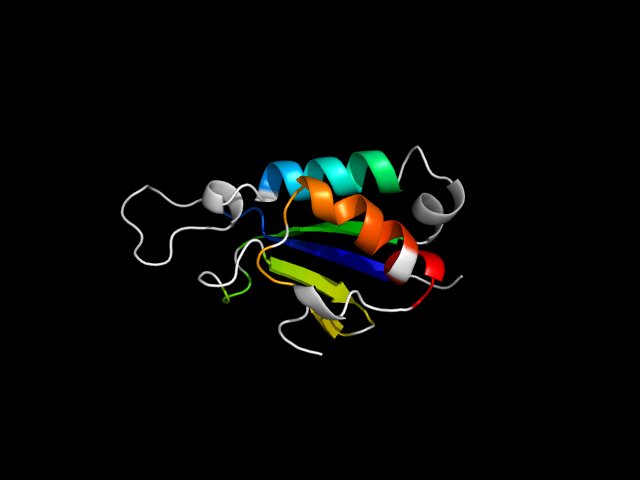 |
d1m2aa_ | A1-A77,A78-A103 | Alpha and beta proteins (a/b) | Thioredoxin fold | Thioredoxin-like | Thioredoxin-like 2Fe-2S ferredoxin | Thioredoxin-like 2Fe-2S ferredoxin
|
Justification for analogy
The motif in 1j22: From the paper for 1j22, in Fig. 2, d1j22a_ have similar catalytic residues as Hjc (d1gefa_). Thus, d1j22a_ IS
an endonuclease superfamily member. Therefore, one strand and one helix in the motif are insertion. In other families, the insertion adopts
different conformations, e.g. d1ckqa_. So this hybrid motif is analogous to thioredoxin.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1j22_Ah ---VKVVVDSR-------------elrSEVVKRLKLLG---------VKLEVKT-LDV----gDYIISE-DVAIERkstPEETAQYIFL-IAKREqee-------
1m2a_Ac aefKHVFVCVQdrppghpqgscaqrgsREVFQAFMEKIqtdpqlfmtTVITPTGcMNAcmmgpVVVVYPdGVWYGQv--KPEDVDEIVEkHLKGGepverlvisk
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1j22_Ah ---VKVVVDSR-------------elrSEVVKRLKLLG---------vKLEVK-TLDV----gDYIIS-EDVAIERkstPEETAQYIFL-IAKREQ-------ee
1m2a_Ac aefKHVFVCVQdrppghpqgscaqrgsREVFQAFMEKIqtdpqlfmttVITPTgCMNAcmmgpVVVVYpDGVWYGQ--vKPEDVDEIVEkHLKGGEpverlvisk
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1j22_Ah ---VKVVVDSR----------EL---RSEVVKRLKLLG---------VKLEVKT-LDV---G-DYIIS-EDVAIERKSTPEETAQYIFL-IAK-RE---------QEE
1m2a_Ac AEFKHVFVCVQDRPPGHPQGSCAQRGSREVFQAFMEKIQTDPQLFMTTVITPTGCMNACMMGPVVVVYPDGVWYGQV-KP-EDVDEIVEKHLKGGEPVERLVISK---
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1j22_Ah ---VKVVVD------------------SRELRSEVVKRLKLLG-----VKLEVKTLDVGD----------YIISE-DVAIERKST------PEETAQYIFLIAKREQEE-----------
1m2a_Ac AEFKHVFVCVQDRPPGHPQGSCAQRGSREVFQ-AFMEKIQTDPQLFMTTVITPTG-----CMNACMMGPVVVVYPDGVWYG----QVKPEDVDEIVEK---HLKG----GEPVERLVISK

