d1jdbc2d1r9ca_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1jdb_Ch
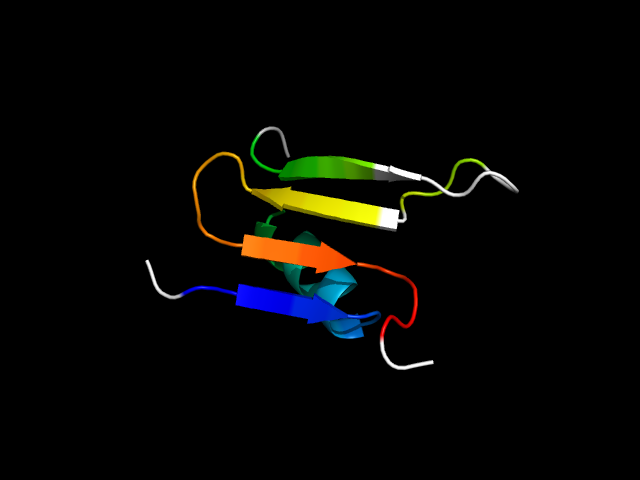 |
d1jdbc2 | C262-C280,C322-C356 | Alpha and beta proteins (a/b) | Flavodoxin-like | Class I glutamine amidotransferase-like | Class I glutamine amidotransferases (GAT) | Carbamoyl phosphate synthetase, small subunit C-terminal domain |
1r9c_Ac
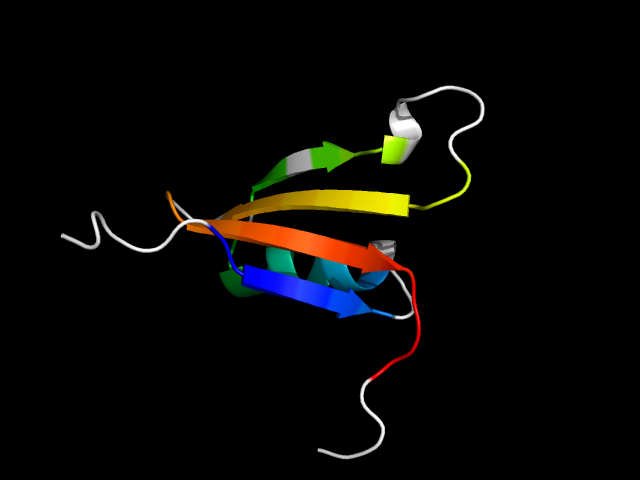 |
d1r9ca_ | A1-A27,A28-A65 | Alpha and beta proteins (a+b) | Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase | Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase | Antibiotic resistance proteins | Fosfomycin resistance protein FosX
|
Justification for analogy
Comparing d1jdbc2 with d1gg9a1 and d1g2ia_, they are homologs (same SCOP superfamily but different family). Note the angle of the helix after strand
4. The motif in 1jdb is a hybrid because two strands in it do not belong to the flavodoxin core.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1jdb_Ch ----diPVFGIC----LGHQLLALASGpaNL-RVThkslfdGTLqGIHRT--DKPAFSFQGHPea--
1r9c_Ac mieglsHMTFIVrdleRMTRILEGVFD-aREvYASdteqfsLSR-EKFFLigDIWVAIMQGEKlaer
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1jdb_Ch ----DIPVFGICLGHQLLALASG---paNLRVTHKsLFDGTLQGIHRTD--KPAFSFQGHPEA--
1r9c_Ac miegLSHMTFIVRDLERMTRILEgvfdaREVYASDtEQFSLSREKFFLIgdIWVAIMQGEKLAer
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1jdb_Ch ----D-IPVF-G--ICLGHQLLALASGPANLRVTH-KS--------LFDGTLQGIHR-TDKPAFSFQGHPE------A
1r9c_Ac MIEGLSHMTFIVRDLERMTRILEGVFD--AREVYASDTEQFSLSRE-------KFFLIGDIWVAIM---QGEKLAER-
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1jdb_Ch ----DIPVFGI----CLGHQLLALASGPAN-LRVTHKSLFDGTLQ------------GIHRTDKPAFSFQGHPEA------
1r9c_Ac MIEGLSHMTFIVRDLERMTRILEGVF----DAREVYA--------SDTEQFSLSREKFFLIGDIWVAIMQG----EKLAER

