d1jnwa_d1ew4a_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1jnw_Ah
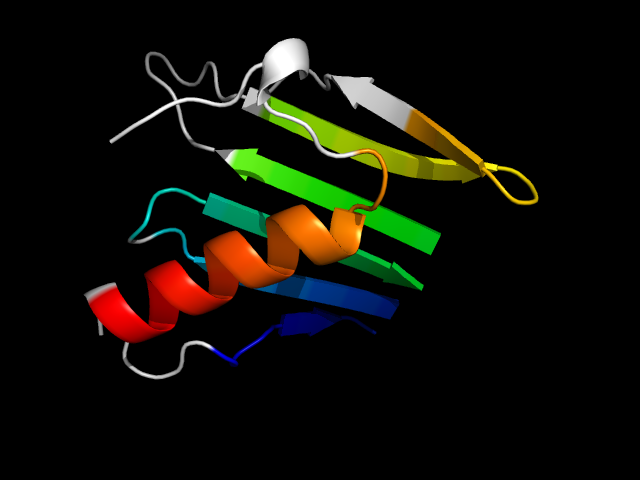 |
d1jnwa_ | A47-A57,A99-A116,A186-A218,A19-A46 | All beta proteins | FMN-binding split barrel | FMN-binding split barrel | PNP-oxidase like | Pyridoxine 5'-phoshate oxidase (PNP oxidase) |
1ew4_Ac
 |
d1ew4a_ | A1-A36,A37-A56,A57-A82,A83-A106 | Alpha and beta proteins (a+b) | N domain of copper amine oxidase-like | Frataxin-like | Frataxin-like | CyaY
|
Justification for analogy
The motif in 1jnw: hybrid motif; compare to 1i0r, the core is a barrel;
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1jnw_Ah -------------------------ladpTAMVVATVSLLFPWHtLERQVMVIG------EQIEFWqggehrlhdRFLYQREN----DAWKidrlapkgglrrrdlpADPLTLFERWLSQACEak----
1ew4_Ac mndsefhrladqlwltieerlddwdgdsdIDCEINGGVLTITFE-NGSKIIINRqeplhqVWLATK---------QGGYHFDLkgdeWICDr-------------sgETFWDLLEQAATQQAGetvsfr
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1jnw_Ah -------------------------LADPTAMVVATVSLLFPWhTLERQVMVIG-----EQIEFWQggehrlhDRFLYQREN-DAWKIdrlapkgglrrrdlpADPLTLFERWLSQ-ACEA---k
1ew4_Ac mndsefhrladqlwltieerlddwdGDSDIDCEINGGVLTITF-ENGSKIIINRqeplhQVWLATK-------QGGYHFDLKgDEWIC-----------drsgETFWDLLEQAATQqAGETvsfr
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1jnw_Ah ---------------------LADPTAMVVAT---VSLLFPWHTLERQVMVIGE-QIEFWQGGE--HRLHDRFLYQRENDAWKIDRLAP-----------------------------------KGGLRRRDLPADPLTLFERWLSQACEAK
1ew4_Ac MNDSEFHRLADQLWLTIEERL-------DDWDGDSDIDCEIN--GGVLTITFENGSKIIINRQEPLH-QVWLATK--------QGGYHFDLKGDEWICDRSGETFWDLLEQAATQQAGETVSFR----------------------------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1jnw_Ah LADPT------------------------------AMVVATVSLLFPWHTLERQVMVIG-----EQIEFWQGGEHRLH--DRFLYQRENDAWKIDRLAPKGGLRRRDLPA----DPLTLFERWLSQACEAK-------
1ew4_Ac -----MNDSEFHRLADQLWLTIEERLDDWDGDSDIDCEINGGVLTITF-ENGSKIIIN-RQEPLHQVWLAT-------KQGGYHFDLKGDEWICD---------------RSGETFWDLLEQAATQQA---GETVSFR

