d1knza_d1vola1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1knz_Ai
 |
d1knza_ | A7-A58,B7-B58 | Multi-domain proteins (alpha and beta) | NSP3 homodimer | NSP3 homodimer | NSP3 homodimer | NSP3 homodimer |
1vol_Ac
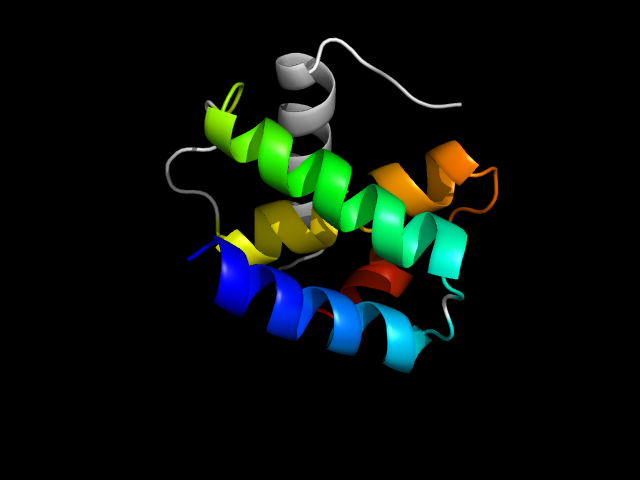 |
d1vola1 | A113-A151,A152-A207 | All alpha proteins | Cyclin-like | Cyclin-like | Transcription factor IIB (TFIIB), core domain | Transcription factor IIB (TFIIB), core domain
|
Justification for analogy
The motif in 1knz: 4-helix bundle uses corresponding parts from the two monomers, so it is an interface motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1knz_Ai tqqmavsIINSSFEAAVVAATSALenmgieydYQDIYSRVKNKFDFVMDDSg-tqqmavsiinSSFEAAVVAATSALENMGIEYDYQDIYSRVKnkfdfvmddsg-----------
1vol_Ac -------AMMNAFKEITTMADRIN-------lPRNKVDRTNNLFRQAYEQKsl-------kgrANDAIASACLYIACRQEGVPRTFKEICAVSRiskkeigrcfklilkaletsvd
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1knz_Ai tqqmavsIINSSFEAAVVAATSALenmgieydYQDIYSRVKNKFDFVMDDSGtqqmavsiiNSSFEAAVVAATSALENMGIEYDYQDIYSRVK-----------nkfdfvmddsg
1vol_Ac -------AMMNAFKEITTMADRIN-------lPRNKVDRTNNLFRQAYEQKS------lkgRANDAIASACLYIACRQEGVPRTFKEICAVSRiskkeigrcfklilkaletsvd
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1knz_Ai TQQMAVSIINSSFEAAVVAATSALENMGIEYDYQDIYSRVKNKFDFVMDDSG--TQQMAVSIINSSFEAAVVAATSALENMGIEYDYQDIYSRVKN---------------------KFDFVMDDSG
1vol_Ac -----A--MMNAFKEITTMADRI-N---L-P-R-NKVDRTNNLFRQAYEQKSLK------G--RANDAIASACLYIACRQEGVPRTFKEICAVSRISKKEIGRCFKLILKALETSVD----------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1knz_Ai TQQMAVSI-INSSFEAAVVAATSALENMGIEYDY---QDIYSRVKNKFDFVMDDSGTQQMAVSIINS------SFEAAVVAATSALENMGIEYDYQDIYSRVKNKFDFVMDDSG-----------------------
1vol_Ac --------AMMNAFKEITTMADRI----------NLPRNKVDRTNNLFRQAYEQK------------SLKGRANDAIASACLYIACRQEGVPRTFKEICAVS------------RISKKEIGRCFKLILKALETSVD

