d1lci__d1lla_1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1lci__h
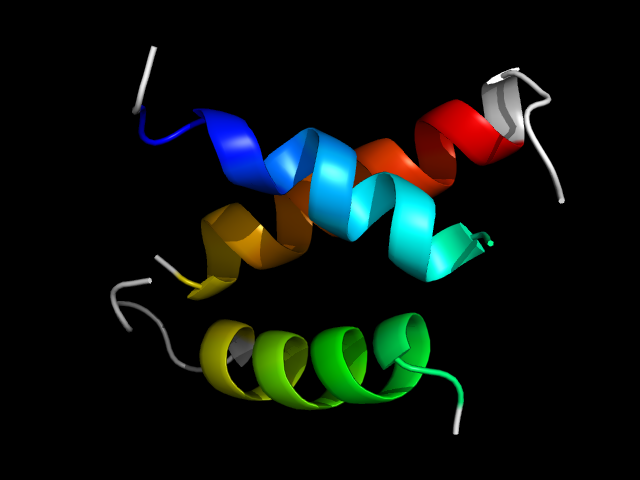 |
d1lci__ | 83-99,20-41,52-74 | Multi-domain proteins (alpha and beta) | Acetyl-CoA synthetase-like | Acetyl-CoA synthetase-like | Acetyl-CoA synthetase-like | Luciferase |
1lla__c
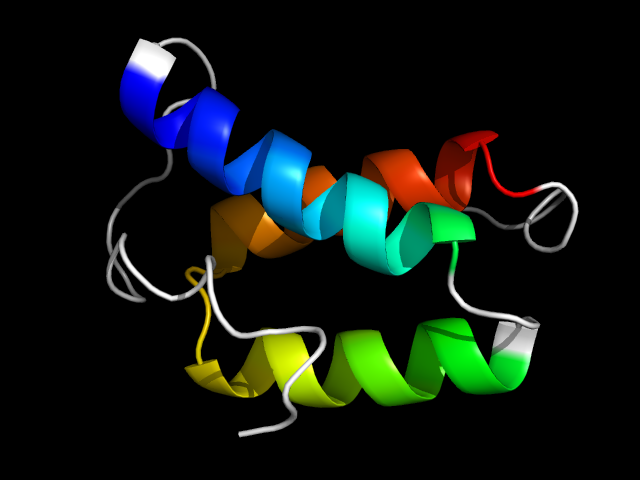 |
d1lla_1 | 30-68,69-84,85-109 | All alpha proteins | Hemocyanin, N-terminal domain | Hemocyanin, N-terminal domain | Hemocyanin, N-terminal domain | Hemocyanin, N-terminal domain
|
Justification for analogy
The motif in 1lci: the first helix is a N-terminal extension, the second one is present in the other duplicate, the third one is a
Rossmann helix. So this is a hybrid motif, but needs circular permutation to make it into a common topology.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lci__h --------------------enSLQFFMPVLGALFIG---gTAGEQLHKAMKRYAlvpgtiatYAEYFEMSVRLAEAMKryglnt---
1lla__c hsdrlknvgklqpgaifscfhpDHLEEARHLYEVFWEagdfNDFIEIAKEARTFV--------NEGLFAFAAEVAVLHRddckglyvp
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lci__h --------------------ENSLQFFMPVLGALFIG---gTAGEQLHKAMKRYAlvpgtiatYAEYFEMSVRLAEAMKRYG---lnt
1lla__c hsdrlknvgklqpgaifscfHPDHLEEARHLYEVFWEagdfNDFIEIAKEARTFV--------NEGLFAFAAEVAVLHRDDCkglyvp
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lci__h --ENSLQFFMPVLGALFIGGTAGEQLHKAMKR--YALV-PGTIATYAEYFEMSVRLAEAMKRYGL--N-------------------------------------T
1lla__c HS------------DRL-----K--NVGK-LQPGAIFSCFH---PDHLEEARHLYEVFWEA--GDFNDFIEIAKEARTFVNEGLFAFAAEVAVLHRDDCKGLYVP-
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lci__h EN----------------------SLQFFMPVLGALFIGGTAG---EQLHKAMKRYALVPGTIATY---AEYFEMSVRLAEAMKRYGLNT---------
1lla__c --HSDRLKNVGKLQPGAIFSCFHPDHLEEARHLYEVFWE----AGDFNDFIEIAKEART-------FVNEGLFAFAAEVAVLHR------DDCKGLYVP

