d1lq7a_d1gqea_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1lq7_Aa
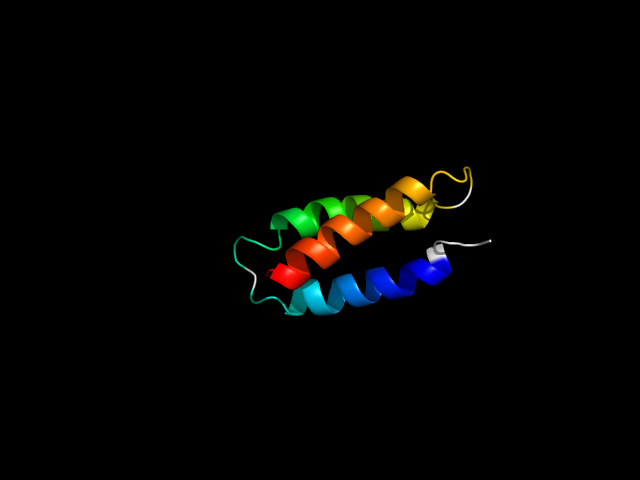 |
d1lq7a_ | A1-A67 | Designed proteins | Designed single chain three-helix bundle | Designed single chain three-helix bundle | Designed single chain three-helix bundle | Alpha3W |
1gqe_Ac
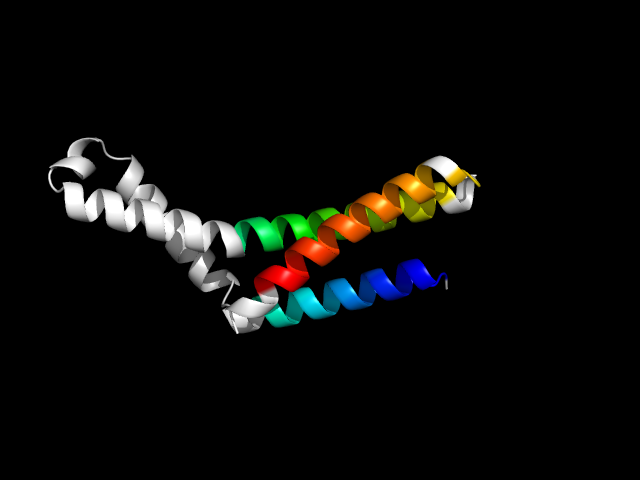 |
d1gqea_ | A4-A117 | Multi-domain proteins (alpha and beta) | Release factor | Release factor | Release factor | Polypeptide chain release factor 2 (RF2)
|
Justification for analogy
1lq7 is an artificial protein, so it is analog to the natural protein 1gqe.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lq7_Aa gsrvKALEEKVKALEEKVKALGg-------------------------------------------GGRIEELKKKWEELKKKIEEL--gGGG-EVKKVEEEVKKLEEEIKKL----
1gqe_Ac ---iNPVNNRIQDLTERSDVLRgyldydakkerleevnaeleqpdvwneperaqalgkerssleavVDTLDQMKQGLEDVSGLLELAveaDDEeTFNEAVAELDALEEKLAQLefrr
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lq7_Aa gsrvKALEEKVKALEEKVKALG-------------------------------------------GGGRIEELKKKWEELKKKIEELG--GGGE-VKKVEEEVKKLEEEIKKL----
1gqe_Ac ---iNPVNNRIQDLTERSDVLRgyldydakkerleevnaeleqpdvwneperaqalgkerssleaVVDTLDQMKQGLEDVSGLLELAVeaDDEEtFNEAVAELDALEEKLAQLefrr
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lq7_Aa GS-RVKALEEKVKALEEKVKALGG--G--------------------------------G-RIEELKKKWEELKKKIEE-LG-G-------GG-E-VKKVEEEVKKLEEEIKKL
1gqe_Ac INPVNNRIQDLTERSDVLRGYLDYDAKKERLEEVNAELEQPDVWNEPERAQALGKERSSLEAVVDTLDQMKQGLEDVSGLLELAVEADDEETFNEAVAELDALEEKLAQLEFRR
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lq7_Aa GS---RVKALEEKVKALEEKVKALGG---------------------------------------GGRIEELKKKWEELKKKIEELGGG---GEVKKVEEEVKKLEEEIKKL----
1gqe_Ac --INPVNNRIQDLTERSDVLRGYLDYDAKKERLEEVNAELEQPDVWNEPERAQALGKERSSLEAVVDTLDQMKQGLEDVSGLLELAVEADDEETFNEAVAELDALEEKLAQLEFRR

