d1lwda_d1a5t_1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1lwd_Ah
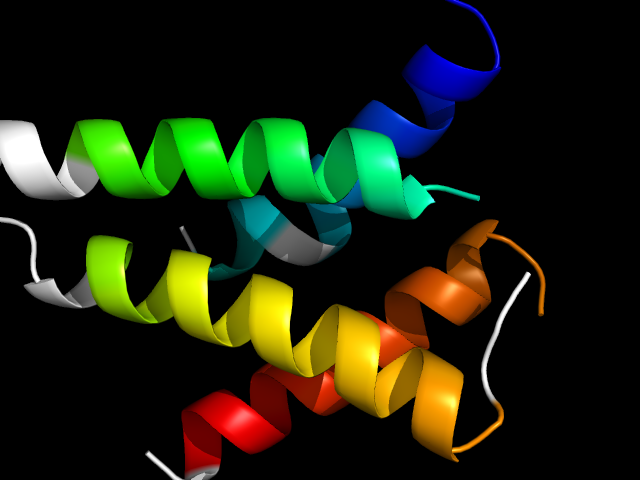 |
d1lwda_ | A17-A36,A328-A373,A393-A413 | Alpha and beta proteins (a/b) | Isocitrate/Isopropylmalate dehydrogenase-like | Isocitrate/Isopropylmalate dehydrogenase-like | Dimeric isocitrate & isopropylmalate dehydrogenases | NADP-dependent isocitrate dehydrogenase |
1a5t__c
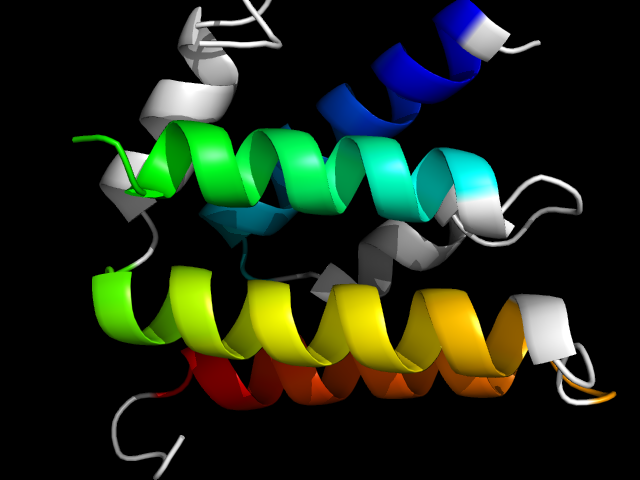 |
d1a5t_1 | 208-237,238-305,306-329 | All alpha proteins | DNA polymerase III clamp loader subunits, C-terminal domain | DNA polymerase III clamp loader subunits, C-terminal domain | DNA polymerase III clamp loader subunits, C-terminal domain | delta prime subunit
|
Justification for analogy
The motif in 1lwd: the 4-helix bundle uses the first and the last helices in the core structure, the other two helices come from
C-terminal extension. So this is a hybrid motif, but the topology is down-down-up-down.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lwd_Ah --DEMTRIIWQFIKEKlILPHv--------------NPIASIFAWTRGLEHRgkldg--------------nqdLIRFAQTLEKVCVETVESGamt--LNTSDFLDTIKSNLDRALGrq---
1a5t__c dnWQARETLCQALAYS-VPSGdwysllaalnheqapARLHWLATLLMDALKR-----vtnvdvpglvaelanhlSPSRLQAILGDVCHIREQLmsvtgINRELLITDLLLRIEHYLQpgvvl
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lwd_Ah --DEMTRIIWQFIKEKlILPH--------------vNPIASIFAWTRGLEHR---------gkldgnqDLIRFAQTLEKVCVETVESG--amtlNTSDFLDTIKSNLDRALGRQ---
1a5t__c dnWQARETLCQALAYS-VPSGdwysllaalnheqapARLHWLATLLMDALKRvtnvdvpglvaelanhLSPSRLQAILGDVCHIREQLmsvtgiNRELLITDLLLRIEHYLQPGvvl
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lwd_Ah DE-MTRIIWQFIKEKLI-LPHV--------------NPIASIFAWTRGLEHRGKL----------D--GN-QDLIRFAQTLEKVC-VET-VE--SGAMTLNTSDFLDTIKSNLDRAL-G--RQ-
1a5t__c DNW-QA-RETLCQA--LAYSVPSGDWYSLLAALNHEQAPARLHWLATLLMDALKRVTNVDVPGLVAELANHLS-PSRLQAILGDVCHIREQLMS-VTGINRE-LLITDLLLRIEH-YLQPGVVL
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1lwd_Ah D---EMTRIIWQFIKEKLILPHVN-----------------PIASIFAWTRGLEHRGKLDGN-------------------QDLIRFAQTLEKVCVETVESGAMTLN-----TSDFLDTIKSNLDRALGRQ-----
1a5t__c -DNWQARETLCQALAYS-------VPSGDWYSLLAALNHEQAPARLHWLATLLMDALKR---VTNVDVPGLVAELANHLSPSRLQAILGDVCHIREQLMS-------VTGINRELLITDLLLRIEHYLQ--PGVVL

