d1miwa2d1gyfa_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1miw_Ah
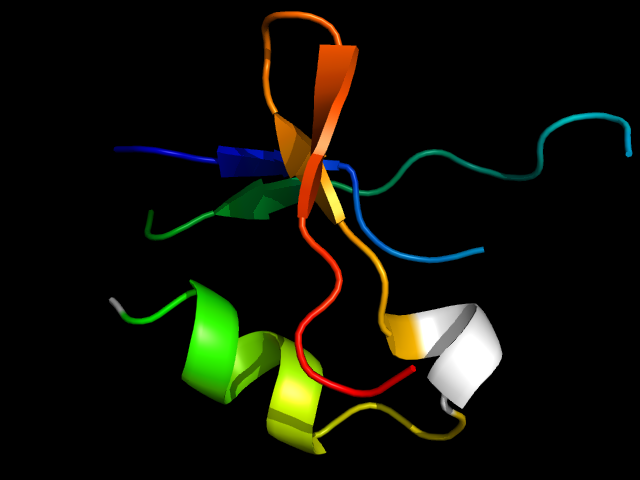 |
d1miwa2 | A20-A27,A36-A47,A102-A131 | Alpha and beta proteins (a+b) | Nucleotidyltransferase | Nucleotidyltransferase | Poly A polymerase head domain-like | tRNA CCA-adding enzyme, head domain |
1gyf_Ac
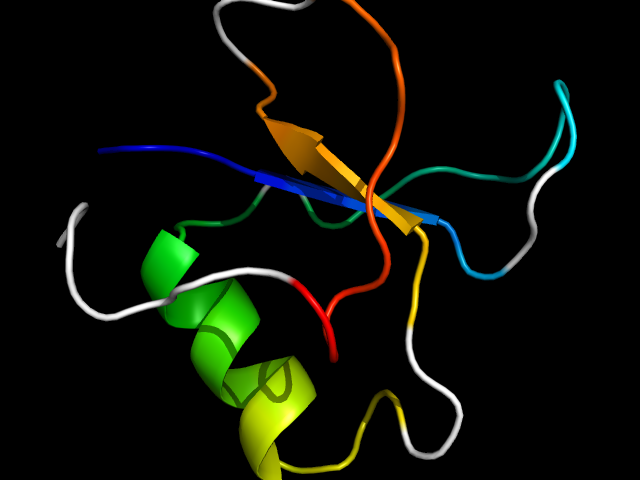 |
d1gyfa_ | A25-A33,A34-A47,A48-A86 | Alpha and beta proteins (a+b) | GYF domain | GYF domain | GYF domain | GYF domain from cd2bp2 protein
|
Justification for analogy
The motif in 1miw: compare 1miw to d1fa0a4 (different family, same superfamily), a C-terminal beta-hairpin is insertion.
The motif in 1gyf: core motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1miw_Ah YDAYFVGG--RPIGDVDI-ATSArSLEEDLKRRDFtmnAIAMDE---YGTIIDPFG-------
1gyf_Ac DVMWEYKWenTGDAELYGpFTSA-QMQTWVSEGYFpdgVYCRKLdppGGQFYNSKRidfdlyt
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1miw_Ah -YDAYFVGGrpIGDVDIATS-aRSLEEDLK--RRDFtmnAIAMD-----eYGTIIDP------fg
1gyf_Ac dVMWEYKWEntGDAELYGPFtsAQMQTWVSegYFPD---GVYCRkldppgGQFYNSKridfdlyt
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1miw_Ah YDAYFVGG--RPIGDVDIATS-ARSLEEDLKR-RDFTMNAI-AMD---EYG-TIIDPF-------G
1gyf_Ac DVMWEYKWENTGDAELYGPFTS-AQMQTWVSEGYFPD--GVYCRKLDPPGGQFYNSKRIDFDLYT-
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1miw_Ah YDAYFVGGRPI------GDVDIATSA-----RSLEEDLKR--RDFTMNA-IAMDE---YGTIIDPFG--------
1gyf_Ac DVMWEYK----WENTGDAELY-----GPFTSAQMQTWVSEGYFPDG---VYCRKLDPPGGQFYNSK-RIDFDLYT

