d1mk5a_d1efub2
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1mk5_Ai
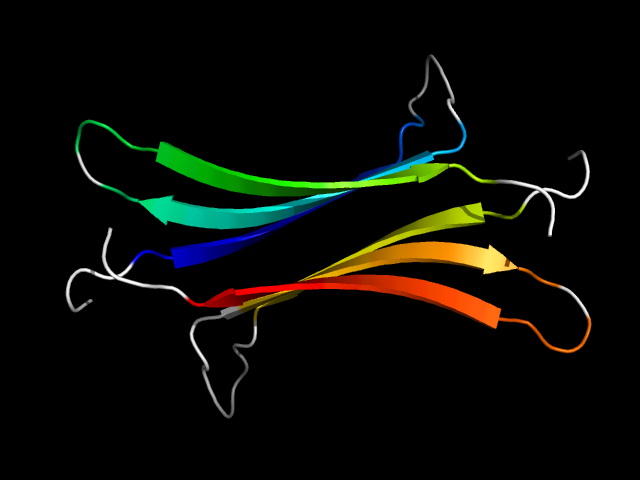 |
d1mk5a_ | A66-A99,A100-A116,B66-B99,B100-B116 | All beta proteins | Streptavidin-like | Avidin/streptavidin | Avidin/streptavidin | Streptavidin |
1efu_Bi
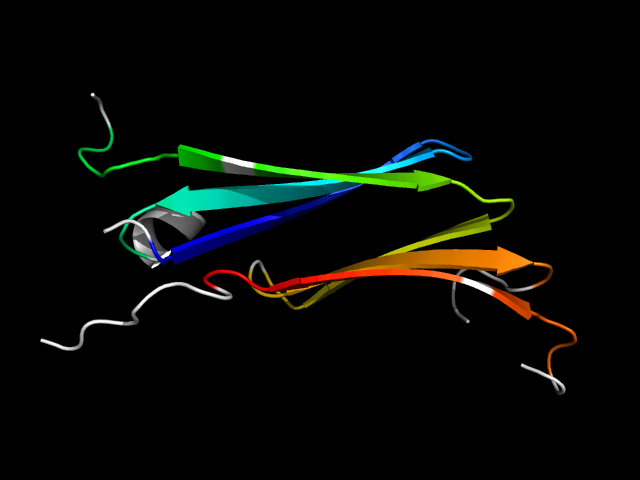 |
d1efub2 | B55-B85,B123-B139,B140-B164,B245-B269 | Alpha and beta proteins (a+b) | EF-Ts domain-like | Elongation factor Ts (EF-Ts), dimerisation domain | Elongation factor Ts (EF-Ts), dimerisation domain | Elongation factor Ts (EF-Ts), dimerisation domain
|
Justification for analogy
The motif in 1mk5: it uses corresponding core elements in two monomers and is an interface motif.
The motif in 1efu: it uses corresponding core elements in two duplicates and is an interface motif.
The two motifs are analogous because 1mk5 is a barrel but 1efu is a a+b structure.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1mk5_Ai tdgsGTALGWTVAWKnnyrnaHSATTWSGQYVGg-------AEARINT-QWLLTSGttetdgsGTALGWTVAwknnyrnahsATTWSGQYVGg-------AEARI-NTQWLLTSgtte----
1efu_Bi --vaADGVIKTKIDG------NYGIILEVNCQTdfvakdakIGENINIrRVAALEG-------DVLGSYQHG---------aRIGVLVAAKGadeellkeHNAEVtGFIRFEVGegiekvet
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1mk5_Ai tdGSGTALGWTVAwknnyrnahsATTWSGQYVGG-----------AEARI-NTQWLLTsgttetdgsgtALGWTVAWKnnyrnaHSATTWSGQYV----gGAEARINTQWLLTSGTTE------
1efu_Bi vaADGVIKTKIDG----------NYGIILEVNCQtdfvakdakigENINIrRVAALEG-----------DVLGSYQHG-----aRIGVLVAAKGAdeellKEHNAEVTGFIRFEVGEGiekvet
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1mk5_Ai TDGSGTALGWTVAWKNNYRNAHSATTWSGQYV---G-----GAEARINTQWLLTSGTTETDGSGTALGWTVAWKNNYRNAHSATTWSGQYVGG-------AEARI-NTQWLLTS--G-TT--E
1efu_Bi --VAADGVIKTKIDG------NYGIILEVNCQTDFVAKDAKIGENINIRRVAALEG-------DVLGSYQHGA---------RIGVLVAAKGADEELLKEHNAEVTGFIRFEVGEGIEKVET-
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1mk5_Ai TDGS----GTALGWTVAWKNNYRNAHSA--TTWSGQYVGGA------------EARIN-TQWLLTSGTTETDGSGTA-LGWTVAWKNNYRN-AHSATTWSGQYVGGAE-----------ARINTQWLLTSGTTE---------
1efu_Bi ----VAADGVIKTKID------------GNYGIILEVNCQ-TDFVAKDAKIGENINIRRVAALEG------------DVLGSYQH------GARIGVLVAAK------GADEELLKEHNAEVT-GFIRFEV---GEGIEKVET

