d1nf2a_d1ptma_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1nf2_Ah
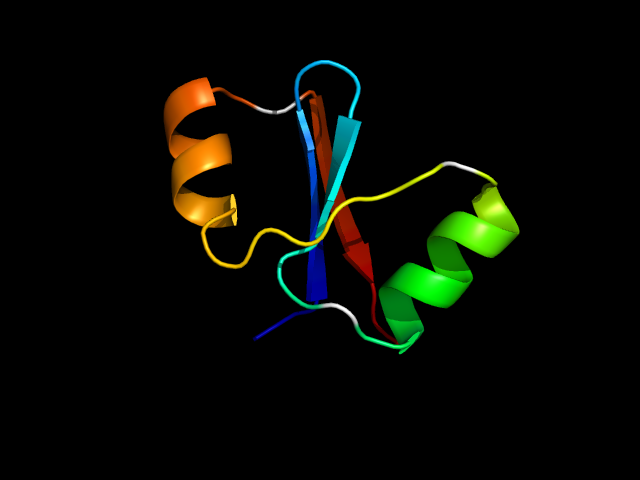 |
d1nf2a_ | A99-A127,A128-A153 | Alpha and beta proteins (a/b) | HAD-like | HAD-like | Predicted hydrolases Cof | Hypothetical protein TM0651 |
1ptm_Ah
 |
d1ptma_ | A146-A202,A258-A289 | Alpha and beta proteins (a/b) | Isocitrate/Isopropylmalate dehydrogenase-like | Isocitrate/Isopropylmalate dehydrogenase-like | PdxA-like | 4-hydroxythreonine-4-phosphate dehydrogenase PdxA
|
Justification for analogy
The motif in 1nf2: compare 1nf2 with 1l6r (same family), the inserted subdomain is a circularly permuted ferredoxin in 1l6r. So the motif in 1nf2 is a hybrid.
The motif in 1ptm: occurs at the interface of the two duplicates, hybrid.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1nf2_Ah --NVHWQAYIDDVLYSEKd--------------------NEEIKSYARHSN------vDYRVEPN---LSELVSKMG--tTKLLLIDT-
1ptm_Ah akKVVMMLATEELRVALAtthlplrdiadaitpallhevIAILHHDLRTKFgiaepraDAVLAMYhdqGLPVLKYQGfgrGVNITLGLp
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1nf2_Ah --nVHWQAYIDDVLYSEK--------------------DNEEIKSYARHSN------vDYRVEP---NLSELVSKMG--tTKLLLIDT-
1ptm_Ah akkVVMMLATEELRVALAtthlplrdiadaitpallheVIAILHHDLRTKFgiaepraDAVLAMyhdQGLPVLKYQGfgrGVNITLGLp
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1nf2_Ah --NVHWQAYIDDVLYSEK--------------------DNEEIKSYARH-S-N--V--DYRVEP---NLSELVSKM--G-TTKLLLIDT-
1ptm_Ah AKKVVMMLATEELRVALATTHLPLRDIADAITPALLHEVIAILHHDLRTKFGIAEPRADAVLAMYHDQG-LPVLKYQGFGRGVNITLGLP
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1nf2_Ah N---VHWQAYIDDVLYSEKDN-------------------EEIKSYARHSNV------------DYRVEPN---LSELVSKMGTT-----KLLLIDT---
1ptm_Ah -AKKVVMMLATEELRVAL---ATTHLPLRDIADAITPALLHEVIAILHHD--LRTKFGIAEPRADAVLAMYHDQGLPVLKYQ---GFGRGVNITL--GLP

