d1nkta3d1ldja1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1nkt_Ah
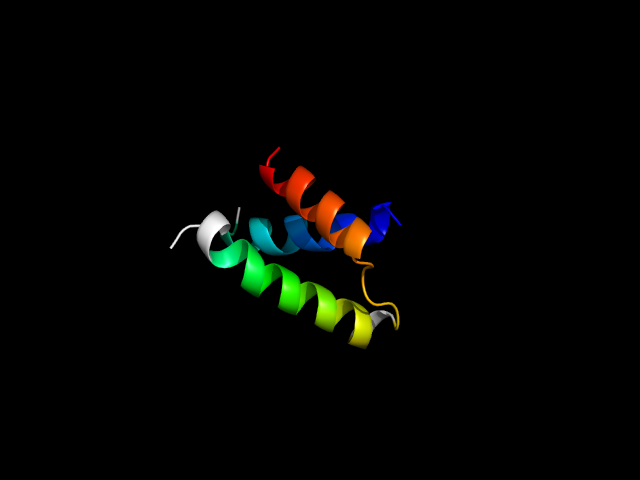 |
d1nkta3 | A106-A123,A57-A96 | Alpha and beta proteins (a/b) | P-loop containing nucleoside triphosphate hydrolases | P-loop containing nucleoside triphosphate hydrolases | Tandem AAA-ATPase domain | Translocation ATPase SecA, nucleotide-binding domains |
1ldj_Ac
 |
d1ldja1 | A687-A724,A725-A776 | All alpha proteins | DNA/RNA-binding 3-helical bundle | "Winged helix" DNA-binding domain | SCF ubiquitin ligase complex WHB domain | Anaphase promoting complex (APC)
|
Justification for analogy
In 1nkt, the 3-helix bundle uses one helix in the N-terminal extra all-alpha subdomain, one helix before the first strand in the
Rossmann-like fold (this helix can be considered as part of the Rossmann fold), and one helix after the P-loop. This is a hybrid motif, but
the topology is up-down-down. So we make a circular permutation.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1nkt_Ah -----------------GKTLTCVLPAYLNALAGn---etlddLLPEAFAVAREAAWRvlDQRPFDVQVMGAAALHLG------------------
1ldj_Ac pmkteqkqeqetthkniEEDRKLLIQAAIVRIMKmrkv----lKHQQLLGEVLTQLSS--RFKPRVPVIKKCIDILIEkeylervdgekdtysyla
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1nkt_Ah --------------------GKTLTC-VLPAYLNALAgnetlddLLPEAFAVAREAAWrvLDQRPFDVQVMGAAALHLG------------------
1ldj_Ac pmkteqkqeqetthknieedRKLLIQaAIVRIMKMRK-----vlKHQQLLGEVLTQLS--SRFKPRVPVIKKCIDILIEkeylervdgekdtysyla
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1nkt_Ah --------------------GKTLTC-VLPAYLNALAGNETLDDLLP-EAFAVAREAAWRVLDQRPFDVQVMGAAA-LHLG------------------
1ldj_Ac PMKTEQKQEQETTHKNIEEDRKLLIQAAI-VRIMKMR---KV--LKHQQLLGEVLTQLS-SR-FKPRVPVIKKCIDIL-IEKEYLERVDGEKDTYSYLA
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1nkt_Ah GKTLTCVLPAYLNALAGNET----------------LDDLLPEAFAVAREAAWRVLDQRPFDVQ-VMGAAALHLG-------------------------------------
1ldj_Ac --------------------PMKTEQKQEQETTHKNIEEDRKLLIQAAIVRIMKMRKV-LKHQQLLGEVLTQLS-SRFKPRVPVIKKCIDILIEKEYLERVDGEKDTYSYLA

