d1o17d2d1jyra_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1o17_Dh
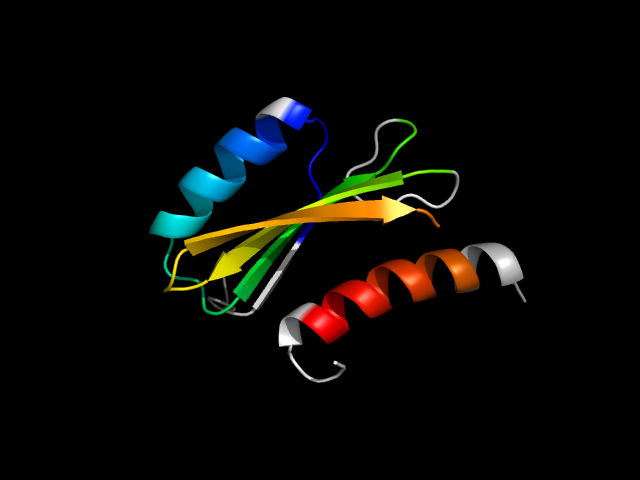 |
d1o17d2 | D182-D248,D280-D303 | Alpha and beta proteins (a/b) | Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain | Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain | Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain | Anthranilate phosphoribosyltransferase (TrpD) |
1jyr_Ac
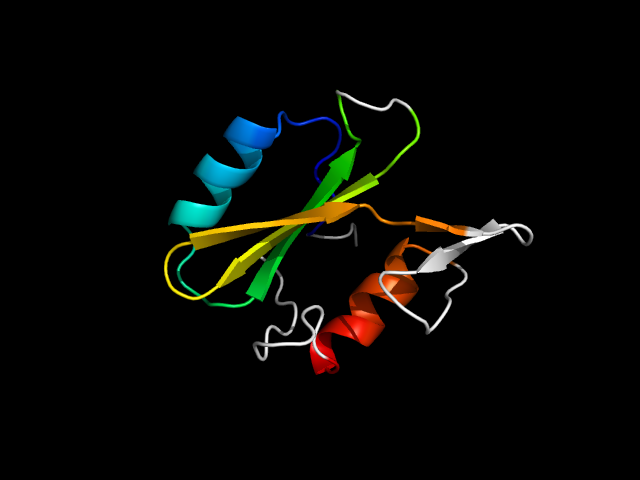 |
d1jyra_ | A56-A120,A121-A151 | Alpha and beta proteins (a+b) | SH2-like | SH2 domain | SH2 domain | Growth factor receptor-bound protein 2 (GRB2)
|
Justification for analogy
The motif in 1o17: compare to d2tpt_2 (same family); two strands are insertions; hybrid motif resulted from a helix-to-strand transition in a Rossmann structure.
The motif in 1jyr: core motif in SH2 fold.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1o17_Dh panakyqlMGVFSKdHLDLLSKSAYELDFNKIILVYGEPgidevspiGNTFMKIVSKRGIEEVKLNV--------kdehvaEFIKINTAVALFaldrvg-------
1jyr_Ac ----gsmaWFFGKI-PRAKAEEMLSKQRHDGAFLIRESEs-----apGDFSLSVKFGNDVQHFKVLRdgagkyflwvvkfnSLNELVDYHRSTsvsrnqqiflrdi
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1o17_Dh panakyqlMGVFSKDhLDLLSKSAYELDFNKIILVYGEpgidevspiGNTFMKIVSKRGIEEVKLNV-----kdehvaeFIKINTAVALFalDRVG------------
1jyr_Ac ----gsmaWFFGKIP-RAKAEEMLSKQRHDGAFLIRES-----esapGDFSLSVKFGNDVQHFKVLRdgagkyflwvvkFNSLNELVDYH--RSTSvsrnqqiflrdi
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1o17_Dh ---PANAKYQLMGVFSKDHLDLLSKSAYELDFNKIILVYGEPG---IDEVSPIGNTFMKIVSKRGIEEVKLNV-------------KDEHVAEFIKINTAVALFALDRVG------------
1jyr_Ac GSM-------AWFFGKIP-RAKAEEMLSKQRHDGAFLIRES-ESAP-------GDFSLSVKFGNDVQHFKVLRDGAGKYFLWVVKF------NS--LNELVDYH-R-STSVSRNQQIFLRDI
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1o17_Dh PANAKYQLMGVF----------SKDHLDLLSKSAYELDFNKIILVYGEPGIDEVSPIG-----NTFMKIVSKRGIEEVKLNVKDEHVA---------------EFIKINTAVALFALDRVG-------------
1jyr_Ac ------------GSMAWFFGKIPRAKAEEMLSKQ---RHDGAFLIRES----------ESAPGDFSLSVKFGNDVQHFKVL-------RDGAGKYFLWVVKFNSLNELVDYHRST------SVSRNQQIFLRDI

