d1ogqa_d1fjgc1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1ogq_Ah
 |
d1ogqa_ | A1-A87 | Alpha and beta proteins (a/b) | Leucine-rich repeat, LRR (right-handed beta-alpha superhelix) | L domain-like | Polygalacturonase inhibiting protein PGIP | Polygalacturonase inhibiting protein PGIP |
1fjg_Cc
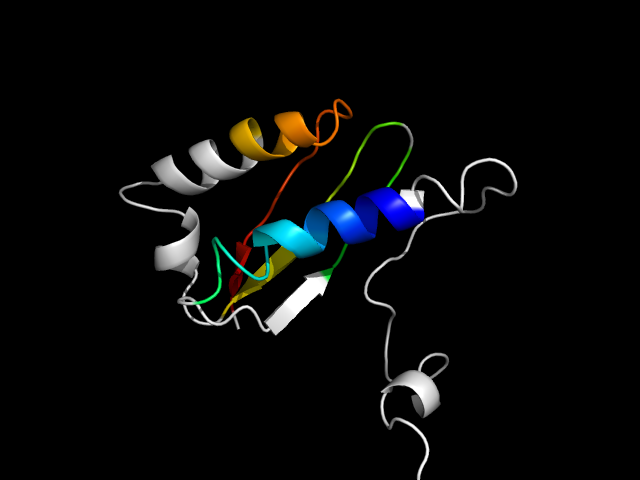 |
d1fjgc1 | C2-C106 | Alpha and beta proteins (a+b) | Alpha-lytic protease prodomain-like | Prokaryotic type KH domain (KH-domain type II) | Prokaryotic type KH domain (KH-domain type II) | Ribosomal protein S3 N-terminal domain
|
Justification for analogy
The motif in 1ogq: compare d1ogqa_ to d1g9ua_ (same superfamily), one strand is insertion. hybrid motif.
The bottom line is that Leucine-rich repeat and KH domain are drastically different structures. So similar motifs come from them are analogs.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ogq_Ah ----------------------------elcNPQDKQALLQIKKDLGnpttlsswlpttdccnrtwlGVLCDtdtqtYRVnNLDLSGlnlpkpy---------piPSSLANLPYLNFLYIGGin
1fjg_Cc gnkihpigfrlgitrdwesrwyagkkqyrhlLLEDQRIRGLLEKELYsagl-------------arvDIERAa----DNV-AVTVHVakpgvvigrggerirvlrEELAKLTGKNVALNVQEv-
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ogq_Ah ------------------------ELCNPQDKQALLQIKKDLG------nPTTLsswlpttdccnrtwlGVLCdtdtqtyrvNNLDLSgLNLP---------------kpyPIPSSLAnlpylnFLYIGGIN-
1fjg_Cc gnkihpigfrlgitrdwesrwyagKKQYRHLLLEDQRIRGLLEkelysagLARV-------------diERAA---------DNVAVT-VHVAkpgvvigrggerirvlreELAKLTG-----kNVALNVQEv
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ogq_Ah ---------------------------EL--CNPQDKQALLQIKKDLGNPTTLSSWLPTTDCCNRTWLGVLCDTDTQTYRVNNLDLSGLNL-PKP-Y----P--IP--SSLA-NLPY-LNFLYIGG-IN
1fjg_Cc GNKIHPIGFRLGITRDWESRWYAGKKQ-YRHLLLEDQRIRGLLEKEL-YSAGLAR------------V-DIER--A-A-DNVAVTVHVAKPG-VVIGRGGERIRVLREE-LAKLTGKNV-ALNVQEV--
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ogq_Ah ELCN----------------------------PQDKQALLQIKKDLGNPTTLSSWLPTTDCCNRTWL------------GVLCDTDTQTYRVN-----NLDLSGLNLPKPYPIPSSL-----------------------ANLPYLN--FLYIGGIN-
1fjg_Cc ----GNKIHPIGFRLGITRDWESRWYAGKKQYRHLLLEDQRIRGLL---------------------EKELYSAGLARVDIER----------AADNVAVTVH--------------VAKPGVVIGRGGERIRVLREELAKLTGK--NVALNVQE--V

