d1ogsa2d1is1a_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1ogs_Ah
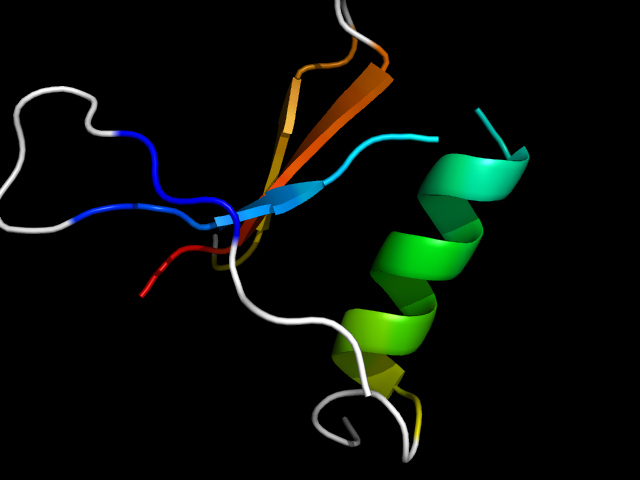 |
d1ogsa2 | A1-A20,A97-A116,A400-A415 | Alpha and beta proteins (a/b) | TIM beta/alpha-barrel | (Trans)glycosidases | beta-glycanases | Glucosylceramidase, catalytic domain |
1is1_Ac
 |
d1is1a_ | A29-A70,A71-A86,A87-A105 | Alpha and beta proteins (a+b) | RRF/tRNA synthetase additional domain-like | Ribosome recycling factor, RRF | Ribosome recycling factor, RRF | Ribosome recycling factor, RRF
|
Justification for analogy
The motif in 1ogs: first two strands come from N-terminal extension, helix belongs to TIM-barrel, last two strands are insertion to TIM-barrel. Hybrid.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ogs_Ah -------------------------arpcIPKsfgyssVVCVCNA--SPPAQNLLLKSYFSeegigy--sPIIVDItkDTFYKQP---
1is1_Ac tgrahpsllsgisveyygaatplnqvanvVAEd----aRTLAITVfdKELTQKVEKAIMMSd-----lglNPMSAG--TIIRVPLppl
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ogs_Ah -------------------------arpcIPKSfgysSVVCVCNA--SPPAQNLLLKSYFseeGIGYsPIIVDItkDTFYKQP---
1is1_Ac tgrahpsllsgisveyygaatplnqvanvVAED----ARTLAITVfdKELTQKVEKAIMM---SDLGlNPMSAG--TIIRVPLppl
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ogs_Ah -----ARPC---------------------IPKSFGYSSVVCVCNA--SPPAQNLLLKSYFSEEGIGY-SPIIVDITKDTFYKQP---
1is1_Ac TGRAH-PSLLSGISVEYYGAATPLNQVANVVAED----ARTLAITVFDKELTQKVEKAIMM---SDLGL-NPMSAG--TIIRVPLPPL
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1ogs_Ah ARP----------------------------CIPKSFGYSSV--VCVCNA-------SPPAQNLLLKSYFSEEGIGYSP-----IIVDITK-DTFYKQP---
1is1_Ac ---TGRAHPSLLSGISVEYYGAATPLNQVANVVAED------ARTLAIT-VFDKELTQKVEKAIMMS------------DLGLNPMSA---GTIIRVPLPPL

