d1oi2a_d1uw1a_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1oi2_Ah
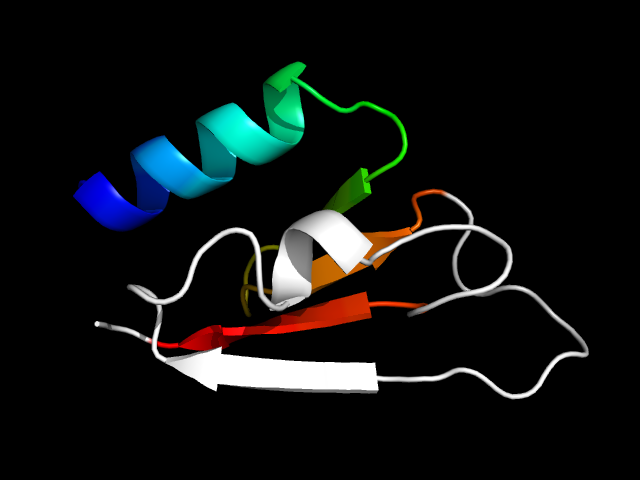 |
d1oi2a_ | A20-A87 | Alpha and beta proteins (a/b) | DAK1/DegV-like | DAK1/DegV-like | DAK1 | Dihydroxyacetone kinase subunit K, DhaK |
1uw1_Aa
 |
d1uw1a_ | A7-A73 | Designed proteins | In vitro evolution products | In vitro evolution products | Function-directed selections | Artificial nucleotide binding protein (ANBP)
|
Justification for analogy
1uw1 is an artificial protein and can be used as analog to natural proteins.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1oi2_Ah -DVLDEQLAGLAKAHP-------SLTLHQD--PVYVTRAdapvagkvallsgggsghepmhcgyigqgmlSGACPGei---
1uw1_Aa dDKKTNWLKRIYRVRPcvkckvaPRNWKVKnkHLRIYNMcktcfnnsidig--------------ddtyhGHDDWLmyads
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1oi2_Ah -DVLDEQLAGLAKAHPS-------LTLHQD--pVYVTRA---DAPV-AGKVallsgggsghepmhcgyigQGMLSGACPGEI---
1uw1_Aa dDKKTNWLKRIYRVRPCvkckvapRNWKVKnkhLRIYNMcktCFNNsIDIG------------------dDTYHGHDDWLMYads
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1oi2_Ah -------------------------D------------VLDEQLAGLAKAHPSLT-LHQDP--------VYVTRADAPVAGKVALLSGGGSGHEPMHCGYIGQGMLSGACPGEI
1uw1_Aa DDKKTNWLKRIYRVRPCVKCKVAPRNWKVKNKHLRIYN-MCKTCFNNSIDIG-DDTYHGHDDWLMYADS---------------------------------------------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1oi2_Ah -DVLDEQLAGLAKAHPS--------LTLHQD---PVYVTRADAPV----------AGKVALLSGGGSGHEPMHCGYIGQGMLS----GACPGEI---
1uw1_Aa DDKKTNWLKRIYRVRP-CVKCKVAPRNWKV-KNKHLRIYN-----MCKTCFNNSIDIGDD-----------------------TYHGHDDWLMYADS

