d1omoa_d1i7b.1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1omo_Ai
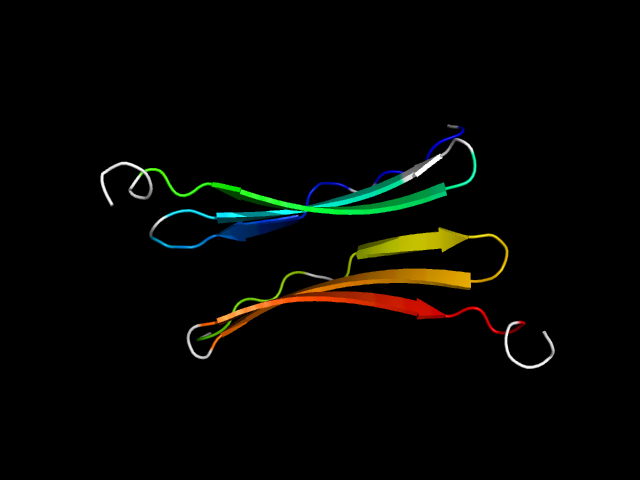 |
d1omoa_ | A34-A47,A48-A59,A60-A76,B34-B47,B48-B59,B60-B76 | Alpha and beta proteins (a/b) | NAD(P)-binding Rossmann-fold domains | NAD(P)-binding Rossmann-fold domains | Ornithine cyclodeaminase-like | Archaeal alanine dehydrogenase |
1i7b_Ai
 |
d1i7b.1 | B4-B23,A103-A119,A153-A166,A171-A185,A276-A289,A316-A328 | Alpha and beta proteins (a+b) | S-adenosylmethionine decarboxylase | S-adenosylmethionine decarboxylase | S-adenosylmethionine decarboxylase | S-adenosylmethionine decarboxylase
|
Justification for analogy
The motif in 1omo: interface motif uses elements from the two dimerization domains;
The motif in 1i7b: interface motif uses elements from the two duplicates;
Since the dimerization domain in 1omo and the duplicate in 1i7b are so different and so unlikely to evolutionarily related, these two motifs are analogs.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1omo_Ai --gKAQMpPKVYLEFE----------kGDLRAMPAhlmGYAGLKWVNSHPgnpdkgKAQMPpKVYLEFE----KGDLRAMPAHLmgYAGLKWVNSHPGnpdk
1i7b_Ai ahfFEGT-EKLLEVWFsrqqpgfdsiqSFFYSRKNfmkNSDCWYLYTLDFpe---sQPDQT-LEILMSEldkpGKFVTTLFVNQsnDYNFVFTSFAKK----
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1omo_Ai --GKAQMPPKvYLEF----------EKGDLRAMPAHLMGYAGLKWVNSHPGNpdkGKAQMPpKVYLEFE----KGDLRAMPAHLMGYAGLKWVNSHPGnpdk
1i7b_Ai ahFFEGTEKL-LEVWfsrqqpgfdsIQSFFYSRKNFMKNSDCWYLYTLDFPE---SQPDQT-LEILMSEldkpGKFVTTLFVNQSNDYNFVFTSFAKK----
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1omo_Ai ---GKAQMPPKVYLEFE--------KGDLRAMPA-HLMGYAGLKWVNSHPGN-PDKGKAQMPPKVYLEFE----KGDLRAMPAHLMGYAGLKWVNSHPGNPDK
1i7b_Ai AHFF-EGTEKLLEVWFSRQQPGFDSIQSFFYSRKNFMKNSDCWYLYTLD-FPE---SQPDQT-LEILMSELDKPGKFVTTLFVNQSNDYNFVFTSFAKK----
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1omo_Ai G-----KAQMPPKVYLEFEKGD---------LRAMPAHLMG------YAGLKWVNSHPGNPDKGK--AQMPPKVYLEFE----KGDLRAMPAHLMGY--AGLKWVNSHPGNPDK-
1i7b_Ai -AHFFEGTEKL-LEVWFSR---QQPGFDSIQSFFYSRKN--FMKNSDCWYLYT-LDFPE------SQPDQT-LEILMSELDKPGKFVTTLFVNQS--NDYNFVFTSFAK-----K

