d1pdz_1d1irum_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1pdz__h
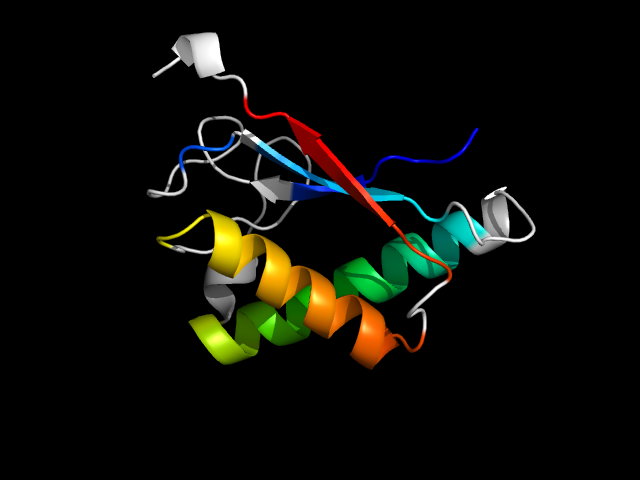 |
d1pdz_1 | 145-250 | Alpha and beta proteins (a/b) | TIM beta/alpha-barrel | Enolase C-terminal domain-like | Enolase | Enolase |
1iru_Mc
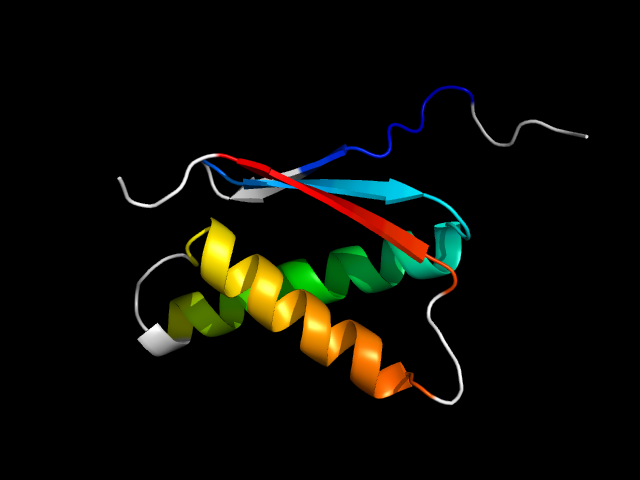 |
d1irum_ | M33-M116 | Alpha and beta proteins (a+b) | Ntn hydrolase-like | N-terminal nucleophile aminohydrolases (Ntn hydrolases) | Proteasome subunits | Proteasome beta subunit (catalytic)
|
Justification for analogy
Compare 1pdz to d1bqg_1 (other family), the anti-parallel strand is the insertion. The motif in 1pdz comes from a TIM-barrel-like structure.
The motif in 1iru_M comes from Ntn-like a+b sandwich. They are analogs.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pdz__h -----PVPAFNVinggshagnklAMqEFMILPtgatsfteAMRMGTEVYHHLKAVIKARFGldatavgdeggfapniLNNKDALDLIQEAIKKAGy-tgKIEIGMDVaasef
1iru_Mc fsihtRDSPKCYkltd-------KT-VIGCSG--------FHGDCLTLTKIIEARLKMYKHsnnk-----------aMTTGAIAAMLSTILYSRRffpyYVYNIIGGldee-
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pdz__h -------PVPAFNVINggshagnklamqEFMILPTgaTSFTEAMRMGTEVYHHLKAVIKARFgldatavgdeggfapnilnnkdALDLIQEAIKKAG-------ytgKIEIGMDVAAsef
1iru_Mc fsihtrdSPKCYKLTD------------KTVIGCS--GFHGDCLTLTKIIEARLKMYKHSNN-------------------kamTTGAIAAMLSTILysrrffpyyvYNIIGGLDEE---
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pdz__h -----P-VPAFNVINGGSHAGNKLAMQEFMILPTGATSFTEAMRMGTEVYHHLKAVIKARFGL-DATAVGDEGGFAPNILNNKDALDLIQEAIKKA---GYTGKIEIGMDV--AASEF
1iru_Mc FSIHTRDSPKCYKLTD----------KTVIGCS--G----FH-GDCLTLTKIIEARLKMYKHSN-N----------KAM-TTGAIAAMLSTILYSRRFFPY-YVYNIIGGLDE----E
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pdz__h P--------VPAFNVINGGSHAGNKLAMQE--FMILPTGATSFTEAMRMGTEVYHHLKAVIKARFGLDATAVGDEGGFAPNIL----NNKDALDLIQEAIKKAGYTGK--------IEIGMDVAASEF
1iru_Mc -FSIHTRDSPKCYKLT--------------DKTVIGCSG--FHGDCLTLTKIIEARLKMYKHSN-------------------NKAMTTGAIAAMLSTILYSR-----RFFPYYVYNIIGGLDEE---

