d1piea1d1q0qa1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1pie_Ah
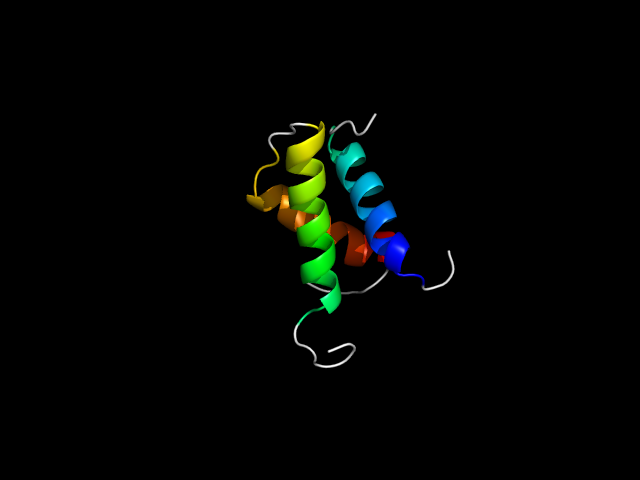 |
d1piea1 | A96-A119,A131-A180 | Alpha and beta proteins (a+b) | Ribosomal protein S5 domain 2-like | Ribosomal protein S5 domain 2-like | GHMP Kinase, N-terminal domain | Galactokinase |
1q0q_Ac
 |
d1q0qa1 | A371-A398,A301-A370 | All alpha proteins | Left-handed superhelix | 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain | 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain | 1-deoxy-D-xylulose-5-phosphate reductoisomerase, C-terminal domain
|
Justification for analogy
The motif in 1pie: two helices come from the "Ribosomal protein S5 domain 2-like" core and the third helix comes from C-terminal
extension. hybrid motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pie_Ah -------dgeLWSNYVKGMIVMLKGAGyeid-------------------------ptasglSSSASLELLVGVVLDDLFnlnVPRLELVQLGQKTENDYIgvnsg
1q0q_Ac epqcvddvlsVDANAREVARKEVMRLAs---klsaltfaapdydrypclklameafeqgqaaTTALNAANEITVAAFLAQq--IRFTDIAALNLSVLEKMDmr---
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pie_Ah ------dGELW-SNYVKGMIVMLKGAG--------------------yeidpTASGL--SSSASLELLVGVVLDDlfNLNVPRLELVQLGQKTENdYIGVnsg
1q0q_Ac epqcvddVLSVdANAREVARKEVMRLAsklsaltfaapdydrypclklameaFEQGQaaTTALNAANEITVAAFL--AQQIRFTDIAALNLSVLE-KMDM--r
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pie_Ah ------DGE--L-WSNYVKGMIVMLKGAGYE-----------------------IDPTASGL-----SSSASLELLVGVVLDDLFNLNVPRLELVQLGQKTENDYIGV-NSG
1q0q_Ac EPQCVD--DVLSVDANAREVARKEVMR-LASKLSALTFAAPDYDRYPCLKLAME-----AFEQGQAATTALNAANEITVAAFL--AQQIRFTDIAALNLSVLEK-MDMR---
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pie_Ah DGEL-----------WSNYVKGMIVMLKGAGYEIDPTASGLS------------------------------SSASLELLVGVVLDDLFNLNVP-----------RLELVQLGQKTENDYIGVNSG---
1q0q_Ac ----EPQCVDDVLSVDANAREVARKEVMRL------------ASKLSALTFAAPDYDRYPCLKLAMEAFEQGQAATTALNAANEITVAA-----FLAQQIRFTDIAALNLSVLEKM----------DMR

