d1pqsa_d1poh__
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1pqs_Aa
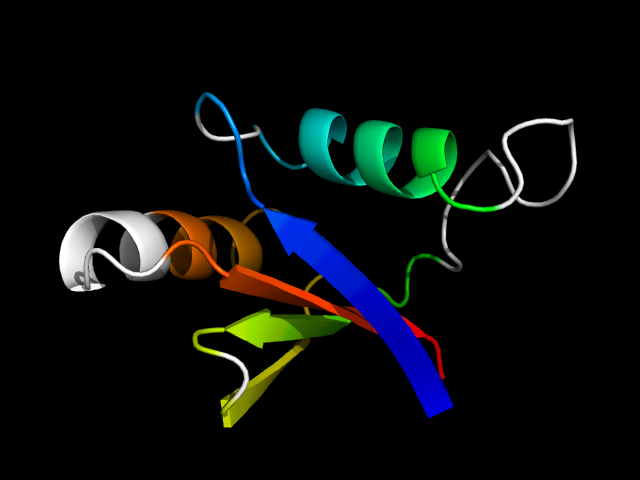 |
d1pqsa_ | A778-A854 | Alpha and beta proteins (a+b) | beta-Grasp (ubiquitin-like) | CAD & PB1 domains | PB1 domain | Cell division control protein 24, CDC24, C-terminal domain |
1poh__c
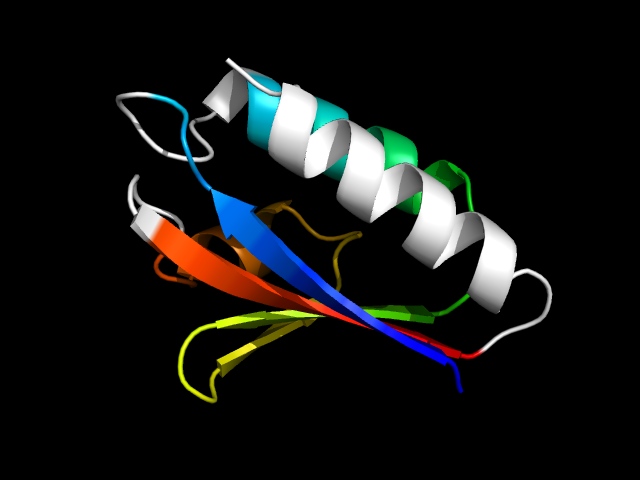 |
d1poh__ | 1-85 | Alpha and beta proteins (a+b) | HPr-like | HPr-like | HPr-like | Histidine-containing phosphocarrier protein (HPr)
|
Justification for analogy
1pqs is a cloning artefact. It should be a ubiquitin fold.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pqs_Aa SEIFTLLVEKv-----wNFDDLIMAINSKIsnthnnnispiTKIKYQDEdGDFVVLGSDEDWNVAKemlaenneKFLNIRLY-------------------
1poh__c MFQQEVTITApnglhtrPAAQFVKEAKGFT-----------SEITVTSN-GKSASAKSLFKLQTLGltq----gTVVTISAEgedeqkavehlvklmaele
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pqs_Aa SEIFTLLVEK-----vwNFDDLIMainskisnthnnniSPITKIKYQDEdGDFVVLGSDEDWNVakEMLAenneKFLNIRLY-------------------
1poh__c MFQQEVTITApnglhtrPAAQFVK-----------eakGFTSEITVTSN-GKSASAKSLFKLQT--LGLT--qgTVVTISAEgedeqkavehlvklmaele
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pqs_Aa -SEIFTLLVEKVWNFDDLIMAINSKISNTHNNNISPITKIKYQDEDGDFVV----LGSDEDWNVAKEMLAENNEKFLNIRL--------------------Y--------------------------
1poh__c M-------------------------------------------FQQEVTITAPNGLHTRPAAQFVKEAKGFTSEITVTSNGKSASAKSLFKLQTLGLTQGTVVTISAEGEDEQKAVEHLVKLMAELE
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1pqs_Aa SEIFTLLVEK---VWNFDDLIMAINSKISNTHNNNISPIT------KIKYQDEDGDFVVLGSDEDWNVAKEMLAENNEK------FLNIRLY-------------------
1poh__c MFQQEVTIT-APNGLHTRPAAQFVKE--------------AKGFTSEITVTSN-GKSASAKSLFKLQTL----------GLTQGTVVTISAEGEDEQKAVEHLVKLMAELE

