d1qd1b1d1n62c2
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1qd1_Bh
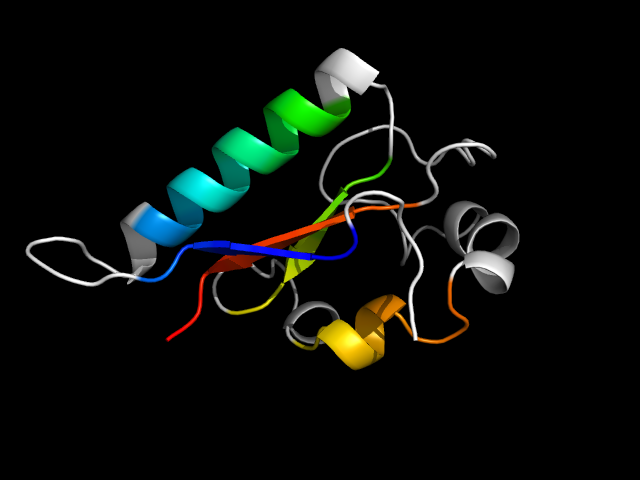 |
d1qd1b1 | B2083-B2180 | Alpha and beta proteins (a+b) | Ferredoxin-like | Formiminotransferase domain of formiminotransferase-cyclodeaminase. | Formiminotransferase domain of formiminotransferase-cyclodeaminase. | Formiminotransferase domain of formiminotransferase-cyclodeaminase. |
1n62_Cc
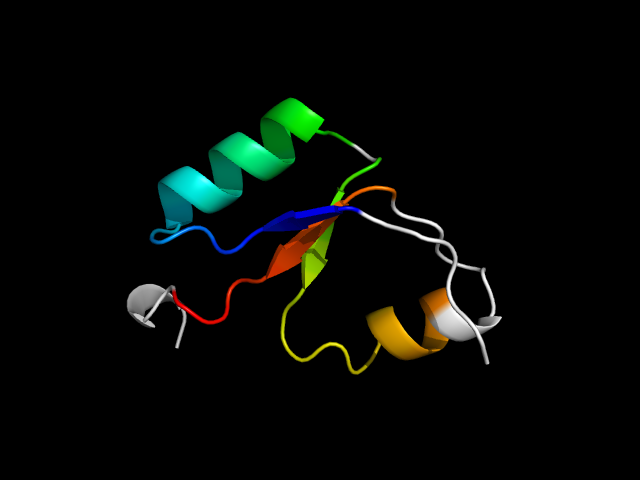 |
d1n62c2 | C1-C61 | Alpha and beta proteins (a+b) | FAD-binding domain | FAD-binding domain | CO dehydrogenase flavoprotein N-terminal domain-like | Carbon monoxide (CO) dehydrogenase flavoprotein, N-terminal domain
|
Justification for analogy
The motif in 1qd1: uses the last strand in ferredoxin and the C-terminal extensions, hybrid. Compare the two duplicates.
The motif in 1n62: stand-along, core motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qd1_Bh prmgalDVCPFIpvrgvtmdeCVRCAQAFGQRLAeelgvPVYLYGeaartagrqSLPALRAGEyealpeklkqaewapdfgpsafvpswgATVAGARK-----
1n62_Cc mipgsfDYHRPK---------SIADAVALLTKLGe----DARPLA---------GGHSLIPIMktrla--------------------tpEHLVDLRDigdlv
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qd1_Bh prmgALDVCPFIpvrgvtmdeCVRCAQAFGQRLAeelgVPVYLYgeaartagrQSLPALRAGEYEAlpeklkqaewapdfgpsafvpswgATVAGARK-----
1n62_Cc mipgSFDYHRPK---------SIADAVALLTKLG----EDARPL---------AGGHSLIPIMKTR--------------------latpEHLVDLRDigdlv
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qd1_Bh -PRMGALDVCPFIPVRGVTMDECVRCAQAFGQRLAEELGVPVYLYGEAARTAGRQSLPALRAGEYEALPEKLKQAEWAPDFGPSAFVPSWGA-TVAGARK-----
1n62_Cc MIPGSFD-YHRPK---------SIADAVALLTKLG----EDARPL---------AGGHSLIPIMKTRL--A-------------------TPEHLVDLRDIGDLV
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qd1_Bh PRMGALD-------VCPFIPVRGVTMDEC-VRCAQAFGQRLAEELGV--PVYLYGEAARTAGRQSLPALRAGEYEALPEKLKQAEWAPDFGPSAFVPSWGA----------TVAGARK------
1n62_Cc -------MIPGSFDYHRPK----------SIADAVALLTKL------GEDARPL---------AGGHSLIP------------------------------IMKTRLATPEHLVDLR-DIGDLV

