d1qh4a2d1ekga_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1qh4_Ah
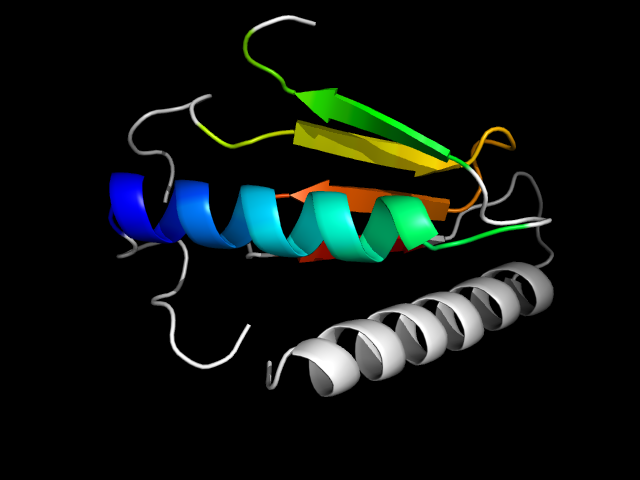 |
d1qh4a2 | A137-A179,A207-A269 | Alpha and beta proteins (a+b) | Glutamine synthetase/guanido kinase | Glutamine synthetase/guanido kinase | Guanido kinase catalytic domain | Creatine kinase, C-terminal domain |
1ekg_Ac
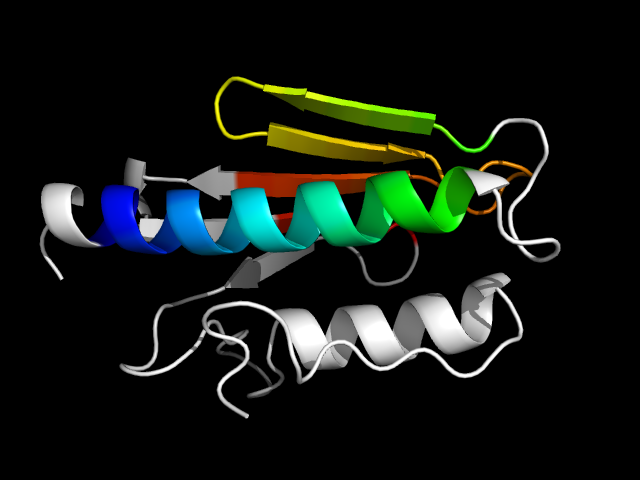 |
d1ekga_ | A90-A129,A130-A208 | Alpha and beta proteins (a+b) | N domain of copper amine oxidase-like | Frataxin-like | Frataxin-like | C-terminal domain of frataxin
|
Justification for analogy
The motif in 1qh4: compare to d2lgsa2 (same superfamily), two strands are insertion; hybrid motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qh4_Ah irgfclpphcSRGERRAIEKLSVEALGSLGgd----lkGKYYALRnmmardwpdARGIWHNDNKTFLVWINee-----dhLRVIsmqkggnmkevftrfctgltqietlfkskny------------------
1ekg_Ac -----ldettYERLAEETLDSLAEFFEDLAdkpytfedYDVSFGS---------GVLTVKLGGDLGTYVINkqtpnkqiwLSSPssgpkrydwtgknwvyshdgvslhellaaeltkalktkldlsslaysgk
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qh4_Ah irgfclpPHCSRGERRAIEKLSVEALGSL--GGDL--kGKYYALrnmmardwpDARGIWHNDNKTFLVWINEE-----DHLRVIS------------mqkggnmkevftrfcTGLT---qieTLFKSK---ny
1ekg_Ac -----ldETTYERLAEETLDSLAEFFEDLadKPYTfedYDVSFG---------SGVLTVKLGGDLGTYVINKQtpnkqIWLSSPSsgpkrydwtgknwvyshdgvslhellaAELTkalktkLDLSSLaysgk
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qh4_Ah IRGFCLPPHCSRGERRAIEKLSVEALGSL--G-----GD--LKGKYYALRNMMARDWPDARGIWHND-N---KTFLVWINE-EDHLRVISMQKGGNMKEVFT-RF-----------------CTGLTQI----------ETLFKS-KN-Y-
1ekg_Ac ----L----DETTYERLAEETLDSLAEFFEDLADKPYTFEDYDVSFGS-------------GVLTVKLGGDLGTYVINKQTPNKQIWLSSP--------SSGP-KRYDWTGKNWVYSHDGVSLHELLAAELTKALKTKL-DL-SSLAYSGK
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qh4_Ah IRGFCLPPHCS------RGERRAIEKLSVEALGSLGGDLK---------GKYYALRNMMARDWPDARGIWHNDNKTFLVWINEEDH----------LRVISMQKGGNMKEVFTRFCTGLTQIETL----------------------------------------FKSKNY-----
1ekg_Ac -----------LDETTYERLAEETLDSLAEFFEDL-----ADKPYTFEDYDVSFG---------SGVLTVKLGGDLGTYVI-----NKQTPNKQIWLSSP-------------------------SSGPKRYDWTGKNWVYSHDGVSLHELLAAELTKALKTKLDLSSL--AYSGK

