d1qnia1d1k4za_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1qni_Ah
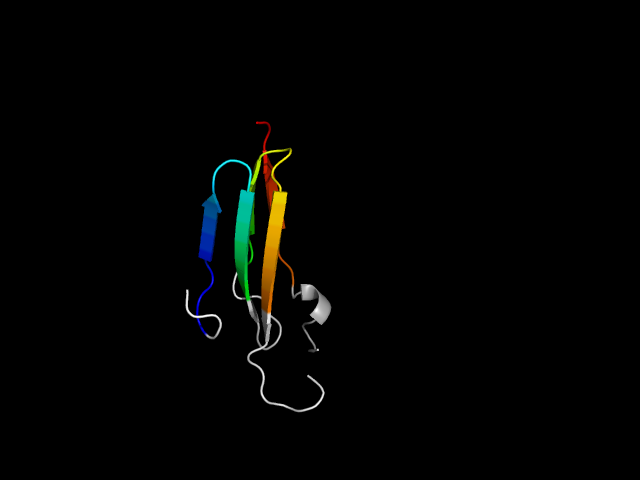 |
d1qnia1 | A472-A524,A565-A581 | All beta proteins | Cupredoxin-like | Cupredoxins | Nitrosocyanin | Nitrous oxide reductase, C-terminal domain |
1k4z_Ac
 |
d1k4za_ | A1368-A1406,A1407-A1524 | All beta proteins | Single-stranded right-handed beta-helix | C-terminal domain of adenylylcyclase associated protein | C-terminal domain of adenylylcyclase associated protein | C-terminal domain of adenylylcyclase associated protein
|
Justification for analogy
The motif in 1qni: compared to 2cbp (same superfamily), the first strand in 1qni is an insertion. So this right-handed beta-helix in 1qni is a
hybrid. Basically, 1qni is a greek-key structure and is fundamentally different from the real beta-helix structures like 1k4z. So the motifs in
1qni and 1k4z are analogs.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qni_Ah vtleSDNKVIRDGNKVRVYMtsvapqygmTDFKVK--EGDEVTVYItnldmvedvchalhmemVGRMLVEAA------------------------------------------------------------------------------------------------------------
1k4z_Ac ----MPPRKELVGNKWFIENye----netESLVIDanKDESIFIGKc---------------sQVLVQIKGKvnaislsetescsvvldssisgmdviksnkfgiqvnhslpqisidksdggniylskeslnteiytscstainvnlpigedddyvefpipeqmkhsfadgkfksavfeh
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qni_Ah vtleSDNKVIRDGNKVRVYMTSVapqygmTDFKVKEGDEVTVYITNLdmvedvchalhmemVGRMLVE----------------------------------------------------------------------------------------------------------aa
1k4z_Ac ----MPPRKELVGNKWFIENYEN--etesLVIDANKDESIFIGKCSQ-------------vLVQIKGKvnaislsetescsvvldssisgmdviksnkfgiqvnhslpqisidksdggniylskeslnteiytscstainvnlpigedddyvefpipeqmkhsfadgkfksavfeh
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qni_Ah --------------------------------------------V-T----------------LESDNK---V-IRDGNKVRVYMTSVAPQYGMTDFKVKEGDEVTVYITNL-DMVEDVCHALHMEMVGRMLVEA----------------------------------------A
1k4z_Ac MPPRKELVGNKWFIENYENETESLVIDANKDESIFIGKCSQVLVQIKGKVNAISLSETESCSVV-LDSSISGMDVIKSNKFGIQVNH-----SLPQISIDKSDGGNIYLSKES----------L--NTEIYTSCSTAINVNLPIGEDDDYVEFPIPEQMKHSFADGKFKSAVFEH-
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1qni_Ah VTLESDNKVIRDGNKVRVYMTSVAPQYGMT---------DFKVKEGDEVTVYITNLDMVEDVCHALHMEM-----VGRMLVEAA-------------------------------------------------------------------------------------------------------------
1k4z_Ac ----MPPRKELVGNKWFIE-----------NYENETESLVIDANKDESIFIG------------------KCSQVLVQIKG---KVNAISLSETESCSVVLDSSISGMDVIKSNKFGIQVNHSLPQISIDKSDGGNIYLSKESLNTEIYTSCSTAINVNLPIGEDDDYVEFPIPEQMKHSFADGKFKSAVFEH

