d1rwha2d1n67a2
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1rwh_Ai
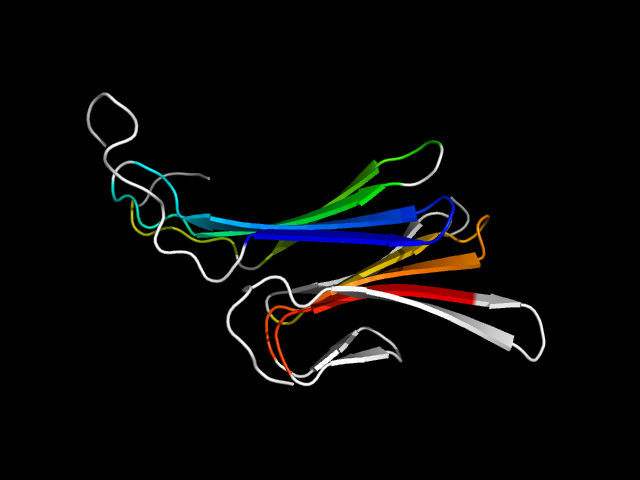 |
d1rwha2 | A462-A526,A642-A709 | All beta proteins | Hyaluronate lyase-like, C-terminal domain | Hyaluronate lyase-like, C-terminal domain | Hyaluronate lyase-like, C-terminal domain | Chondroitinase AC |
1n67_Ah
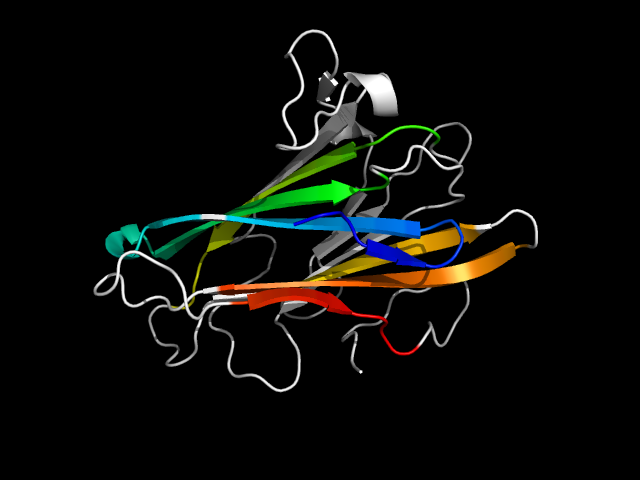 |
d1n67a2 | A370-A499,A500-A560 | All beta proteins | Common fold of diphtheria toxin/transcription factors/cytochrome f | Bacterial adhesins | Fibrinogen-binding domain | Clumping factor A
|
Justification for analogy
The motif in 1rwh: This protein has two all-beta domains: Hyaluronate lyase-like (H) and Supersandwich (S). All proteins containing H have
S, but not vice versa. Since H and S are two separate domains, this motif is an interface motif, using some strands in H and some in S.
The motif in 1n67: uses immunoglobulin core elements as well as a C-terminal extension, hybrid.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1rwh_Ai egqwgaavpadewSGATALGEVAAVGQH-LVGPgRTGLTARKSWFVsg--------------------------------------------------------------------------------DVTVCLGAdISTASgakvegnkysvirndaTAQSVEFK---TAKTTAATFWKpgmag--dlgasGPACVVFSrhgnelslavseptqkaag
1n67_Ah -------------EKYGKFYNLSIKGTIdQIDK-TNNTYRQTIYVNpsgdnviapvltgnlkpntdsnalidqqntsikvykvdnaadlsesyfvnpenfedvtnsvnitfpnpnqykvefntpddqiTTPYIVVV-NGHIDpns--------kgdlaLRSTLYGYnsnIIWRSMSWDNEvafnngsgsgdgIDKPVVPEqpdepgeiepipek-----
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1rwh_Ai egqwgaavpadewsgaTALG----EVAAVG-QHLVGpgrtgLTARKSWFVSGDvtvclgadistasgakveGNKYSVIR-----------------------------------------------------------------------------------ndATAQSVEFKT-------aKTTAATFWKpgmagdlgaSGPACVVFS-RHGNelslavseptqkaag
1n67_Ah ---------------eKYGKfynlSIKGTIdQIDKT-----NNTYRQTIYVNP------------sgdnviAPVLTGNLkpntdsnalidqqntsikvykvdnaadlsesyfvnpenfedvtnsvnitfpnpnqykvefntpddqittpyivvvnghidpnskgDLALRSTLYGynsniiwrSMSWDNEVA--fnngsgsGDGIDKPVVpEQPD----epgeiepipek
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1rwh_Ai ---EGQWGAAVPADEWSGATALGEVAAVGQHLVGPGRTGLTARKSWFV-SGD--------------------------VTVCLGADISTASGAKVEGNKYSVIRNDATAQSVEFKTA----------------KTTAATFWKPGMAGDLGASG-----PACVVFSRHGNELSL-------------------------AV---------S---------------------------------EPTQK--AAG
1n67_Ah EKY---------------GKFYNLSIKGTIDQID--KTNNTYRQTIYVNPSGDNVIAPVLTGNLKPNTDSNALIDQQN----------------------------TSIKVYKVDNAADLSESYFVNPENFED-----------VTNSVNITFPNPNQYKVEFNTP---DDQITTPYIVVVNGHIDPNSKGDLALRSTLYGYNSNIIWRSMSWDNEVAFNNGSGSGDGIDKPVVPEQPDEPGEIEPIPEK---
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1rwh_Ai EGQWGAAVPADEWSGATALGEVAAVGQHL-VGPGRTGLTARKSWFVSG-----------------------------------------------------------------------------------DVTVCLGADISTASGAKVEGNKYSVIRNDAT-------AQSVEFKT----AKTTAATFWKPGMAGDLGASG--------------PACVVFSRHGNELSLAVSEPTQKAAG---------------
1n67_Ah -------------EKYGKFYNLSIKGTIDQIDKT-NNTYRQTIYV---NPSGDNVIAPVLTGNLKPNTDSNALIDQQNTSIKVYKVDNAADLSESYFVNPENFEDVTNSVNITFPNPNQYKVEFNTPDDQITTPYIVVVNGHIDPN----------------SKGDLALRSTLYGY-NSNIIWRSMSWDN------------EVAFNNGSGSGDGIDKPVVP--------------------EQPDEPGEIEPIPEK

