d1smta_d1b7ta4
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1smt_Ai
 |
d1smta_ | A28-A59,A101-A121,B28-B59,B101-B120 | All alpha proteins | DNA/RNA-binding 3-helical bundle | "Winged helix" DNA-binding domain | ArsR-like transcriptional regulators | SmtB repressor |
1b7t_Ac
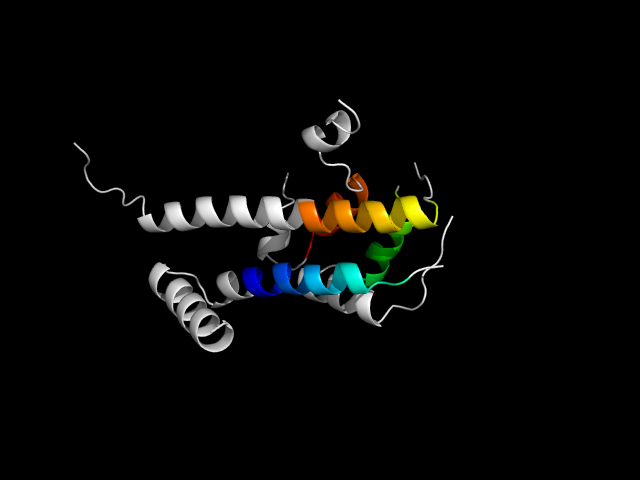 |
d1b7ta4 | A323-A364,A371-A402,A410-453,A591-A624 | Alpha and beta proteins (a/b) | P-loop containing nucleoside triphosphate hydrolases | P-loop containing nucleoside triphosphate hydrolases | Motor proteins | Myosin S1, motor domain
|
Justification for analogy
The motif in 1smt: it comes from a dimer interface, so it is an interface motif.
As long as the DALI hits are not DNA/RNA-binding 3-helix bundle, they can be used as analogs to the interface motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1smt_Ai -----------------------iAPEVAQSLAEFFAVLAdpnrlrllsllarse--------------qlqdhHIVALYQNALDHlqec-iapeVAQSLAEFFAVLADpnrlrllsllarse--------------------qlqdHHIVALYQNAldhlqe-----
1b7t_Ac ddveefklcdeafdilgftkeekqSMFKCTASILHMGEMKfk-------------qaesdgtaeaekvaflcgiNAGDLLKALLKPk---vtkgqNMNQVVNSVGALAKslydrmfnwlvrrvnktldtkakrnitgwleknkdpinENVVALLGASkeplvaelfka
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1smt_Ai -----------------------iAPEVAQSLAEFFAVLaDPNR--LRLLsllarseqlqdhHIVALYQNALDHLQECiapeVAQSLAEFFAVLADpnRLRLLSLL----------------------arseqlqdHHIVALYQnalDHLQ-------e
1b7t_Ac ddveefklcdeafdilgftkeekqSMFKCTASILHMGEM-KFKQaeSDGTaeaekvaflcgiNAGDLLKALLKPKVTK--gqNMNQVVNSVGALAK--SLYDRMFNwlvrrvnktldtkakrnitgwleknkdpinENVVALLG--aSKEPlvaelfka
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1smt_Ai ---------------------I--APEVAQSLAEFFAVLADPN-RLRLLSLL--ARSEQLQ---D---HHIVALYQNA-LDH-LQECIAPEVAQSLAEFFAVLAD-PNRLRLLS---------------------------LLARSEQLQDHHIVALY--QNAL---------DHLQE
1b7t_Ac DDVEEFKLCDEAFDILGFTKEEKQSMFKCTASILHMGEM-KFKQAESDG--TAE-----AEKVAFLCGINAGDLLKALLKPKVTKG----QNMNQVVNSVGALAKSLYDRMFNWLVRRVNKTLDTKAKRNITGWLEKNKDP-------INENVVAL-LGASK-EPLVAELFKA-----
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1smt_Ai IAP-------------------EVAQSLAEFFAVLADPNRLRLLSLLARSEQLQDHHIVAL-----------------------YQNALDHLQECIA------------PEVAQSLAEFFAVLADPNRLRLLSLLARSEQLQDHHIVALYQNALDHLQE-----------------------------------------
1b7t_Ac ---DDVEEFKLCDEAFDILGFTKEEKQSMFKCTASIL--HMGEM-----------------KFKQAESDGTAEAEKVAFLCGINAGDLLKALLK---PKVTKGQNMNQVVNSVGALAKSLYDRM--FNWLVRRVNKTL---------------------DTKAKRNITGWLEKNKDPINENVVALLGASKEPLVAELFKA

