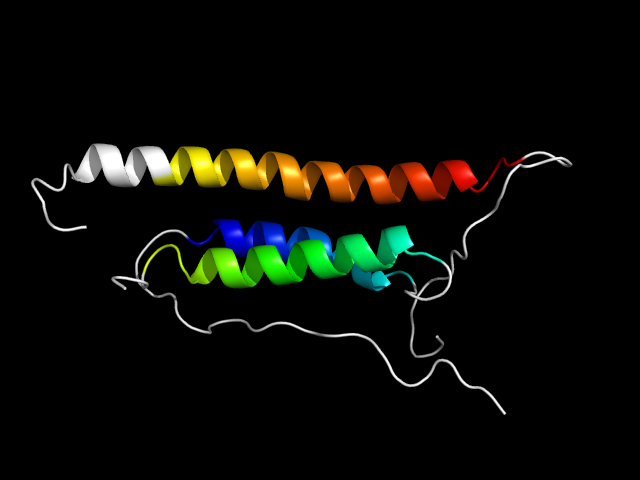
Justification for analogy
The motif in 1st6: comparing the 8 duplicates, we know that the helix before the last duplicate is an insertion. So the three helix bundle
is a hybrid motif with one inserted helix and two core helices.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1st6_Ah ------------------------ppppeekdeefpeqkageainQPMMMAARQLHDEARKWss------kgNDIIAAAKRMALLMAEMSRLVRggsggrtnisdeeseqaTEMLVHNAQNLMQSVKETVREAEAASIKIrtdagftlrwvrktpwy
1r5i_Dc mklrvenpkkaqkhfvqnlnnvvftnkelediynlsnkeetkevlKLFKLKVNQFYRHAFGIvndynglleyKEIFNMMFLKLSVVFDTQRKEA----------------nNVEQIKRNIAILDEIMAKADNDLSYFISQnknf-------------
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1st6_Ah -------------ppppeeKDEEF----PEQK--AGEA-----iNQPMMMAARQLHDEARKWS------skgNDIIAAAKRMALLMAEMSRLVRggsggrtnisdeeseqaTEMLVHNAQNLMQSVKETVREAEAAS-IKIRTdagftlrwvrktpwy
1r5i_Dc mklrvenpkkaqkhfvqnlNNVVFtnkeLEDIynLSNKeetkevLKLFKLKVNQFYRHAFGIVndynglleyKEIFNMMFLKLSVVFDTQRKEA----------------nNVEQIKRNIAILDEIMAKADNDLSYFiSQNKN--------------f
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1st6_Ah ----------------PPPP-EEKD-EEFPE-QK---A-GE-A--IN-Q-PMMMAARQLHDEARK-W---S--SKG-NDIIAAAKRMALLMAE-MSRLVRGGSGGRTNISDEESEQATEMLVHNAQNLMQSVKETVREAEAASIKIRT-DAGFTLRWVRKTPWY
1r5i_Dc MKLRVENPKKAQKHFV---QN-LNNVVFTNKELEDIYNLSNKEETKEVLKLFKLKVNQFYRHAFGIVNDYNGL-LEYKEIFNMMFLKLSVVFDTQRKEAN-----------------NVEQIKRNIAILDEIMAKADNDLSYFISQNKN-------F-------
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1st6_Ah PPPPEEKDEEF------------------------PEQKAGEAINQ-------------------------PMMMAARQLHDEARKWSSKGNDIIAAAKRMALLMAEMSRLVRGGSGGRTNISD-----EESEQATEMLVHNAQNLMQSVKETVREAEAASIKIRTDAGFTLRWVRKTPWY--
1r5i_Dc -----------MKLRVENPKKAQKHFVQNLNNVVFTNKE-------LEDIYNLSNKEETKEVLKLFKLKVNQFYRHAFGIVNDYNGLL-----EYKEIFNMMFLKLSVVFDTQR----------KEANNVEQIKRNIAILDEIMAKADNDLSYFISQNK----------------------NF