d1tv8a_d1nbwb_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1tv8_Ah
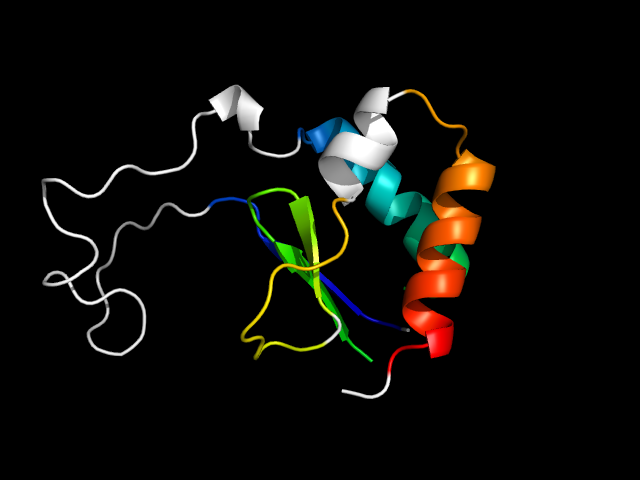 |
d1tv8a_ | A13-A67,A265-A314 | Alpha and beta proteins (a/b) | TIM beta/alpha-barrel | Radical SAM enzymes | MoCo biosynthesis proteins | Molybdenum cofactor biosynthesis protein A MoaA |
1nbw_Bc
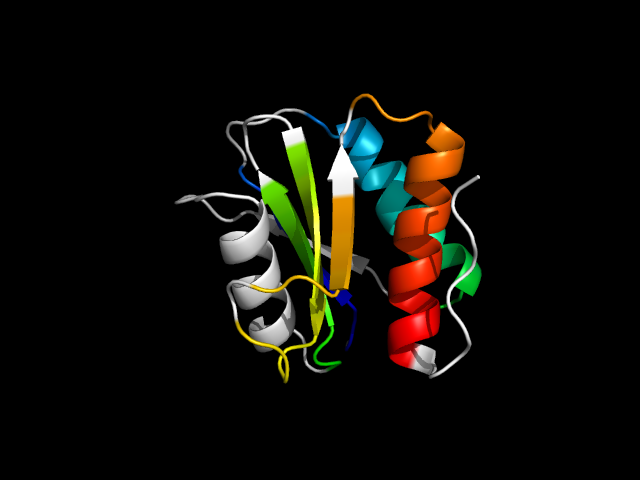 |
d1nbwb_ | B5-B58,B59-B117 | Alpha and beta proteins (a/b) | Anticodon-binding domain-like | B12-dependent dehydatase associated subunit | Glycerol dehydratase reactivase, beta subunit | Glycerol dehydratase reactivase, beta subunit
|
Justification for analogy
The motif in 1tv8: a hybrid motif in an opened TIM-barrel.
1nbw has basically a mini-Rossmann core.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1tv8_Ah iRDLRLSVTdrcnfrcdycmpkevfgddfvflpknellTFDEMARIAKVYAELGV-------------------------TRARLSS----DGKFYgCLFAT--VDGFNvkafirsGVTDEELKEQFKALWQIRdd-------
1nbw_Bc pPGVRLFYDpr-------------------------ghHAGAINELCWGLEEQGVpcqtitydgggdaaalgalaarsspLRVGIGLsasgEIALT-HAQLPadAPLATgh----vTDSDDQLRTLGANAGQLVkvlplsern
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1tv8_Ah irDLRLSVTdrcnfrcdycmpkevfgddfvflpKNELlTFDEMARIAKVYAELGV-------------------------TRARLSSD----GKFYGCLF-ATVDGFNVKafIRSGVTD-eelkeqFKALWQI----rdd
1nbw_Bc ppGVRLFYD------------------------PRGH-HAGAINELCWGLEEQGVpcqtitydgggdaaalgalaarsspLRVGIGLSasgeIALTHAQLpADAPLATGH--VTDSDDQlrtlganAGQLVKVlplsern
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1tv8_Ah -IRDLRLSVTDRCNFRCDYCMPKEVFGDDFVFLPKNELLTFDEMARIAKVYAELGV-------------------------TRARLSSD----GKFYGCLFATVDG-FNVKAFIRSGVTDEELKEQFKALW---QIR----DD
1nbw_Bc P-PGVRLFYD-P----------------------R--GHHAGAINELCWGLEEQGVPCQTITYDGGGDAAALGALAARSSPLRVGIGLSASGEIALTHAQLPADAPLATGHV-TD-SDDQLRTLGANAGQLVKVLPLSERN--
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1tv8_Ah IR----DLRLSVTDRCNFRCDYCMPKEVFGDDFVFLPKNELLT----FDEMARIAKVYAELGVT-----------------------------RARLSSDGKFYG-CLFATVDGFNVKAFIRSG------------VTDEELKEQFKALWQIRDD----------
1nbw_Bc --PPGVRLFYDP-------------------------------RGHHAGAINELCWGLEEQG--VPCQTITYDGGGDAAALGALAARSSPLRVGIGLSASGEIALTHAQL--------------PADAPLATGHVTDSDDQLRTLGANAGQL---VKVLPLSERN

