d1uw1a_d1q9ua_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1uw1_Aa
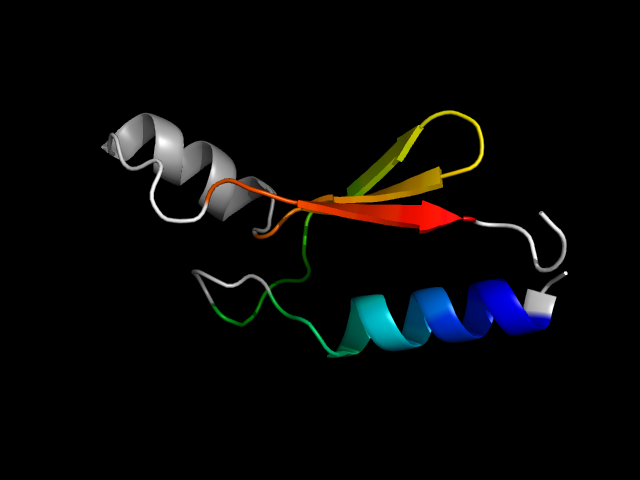 |
d1uw1a_ | A7-A73 | Designed proteins | In vitro evolution products | In vitro evolution products | Function-directed selections | Artificial nucleotide binding protein (ANBP) |
1q9u_Ac
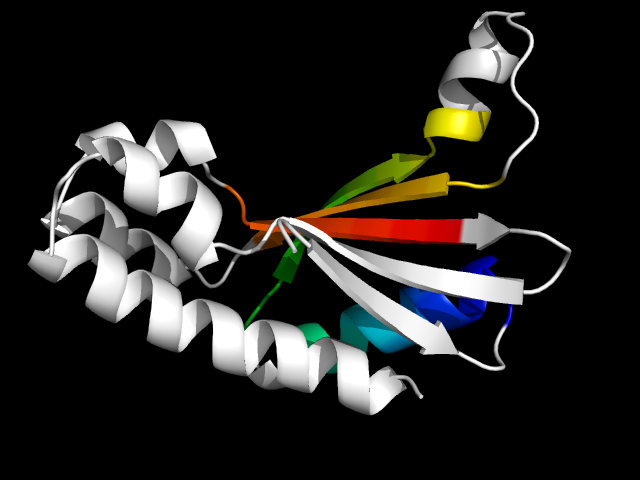 |
d1q9ua_ | A1-A127 | Alpha and beta proteins (a+b) | TBP-like | TT1751-like | TT1751-like | Unnamed hypothetical protein
|
Justification for analogy
1uw1 is an artificial protein, so it is analogous to the natural protein 1q9u.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1uw1_Aa --------ddKKTNWLKRIYRVRPcvkcKVAPRNWKVKN------------KHLRIYNmcktcfnnsidig---ddtyHGHDDWLmyads---------------------------------------
1q9u_Ac mfhytvdvstGMNETIERLEESLKq--eGFGVLWQFSVTeklqekgldfstPMVILEVcnpqeaarvlnenllvgyflPCKLVVYqengttkigmpkptmlvgmmndpalkeiaadiekrlaacldrcr
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1uw1_Aa --------DDKKTNWLKRIYRVRPcvkcKVAPRN-WKVKN------------KHLRIYNMCKTCFNNSIDIG--DDTYH-GHDDWLmYADS----------------------------------------
1q9u_Ac mfhytvdvSTGMNETIERLEESLK---qEGFGVLwQFSVTeklqekgldfstPMVILEVCNPQEAARVLNENllVGYFLpCKLVVY-QENGttkigmpkptmlvgmmndpalkeiaadiekrlaacldrcr
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1uw1_Aa ---------DDKK-TNWLKRIYRVRP-CVKCKVAPR-NWKVKN------------KHLRIYNMCKTC---FNNS------IDIG-DDTYH-GHDDWLMYADS---------------------------------------
1q9u_Ac MFHYTVDVS--TGMNETIERLEESLKQ-E--GF-GVLWQFSVTEKLQEKGLDFSTPMVILEVCN--PQEA---ARVLNEN---LLVGYFLPCKLVVYQENGTTKIGMPKPTMLVGMMNDPALKEIAADIEKRLAACLDRCR
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1uw1_Aa D----------------DKKTNWLKRIYRVRPCVKCKVAPRNWKVKNK--------------------------HLRIYNMCKTCFNNSIDIGDDTYHG---HDDWLMYADS--------------------------------------------
1q9u_Ac -MFHYTVDVSTGMNETIERLEESLKQE---------------------GFGVLWQFSVTEKLQEKGLDFSTPMVILEVC--NPQEAARVLNE-NLLVG-YFLPCKLVVY---QENGTTKIGMPKPTMLVGMMNDPALKEIAADIEKRLAACLDRCR

