d1vaoa2d1no5a_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
1vao_Ah
 |
d1vaoa2 | A6-A41,A71-A95,A117-A123 | Alpha and beta proteins (a+b) | FAD-binding domain | FAD-binding domain | FAD-linked oxidases, N-terminal domain | Vanillyl-alcohol oxidase |
1no5_Ac
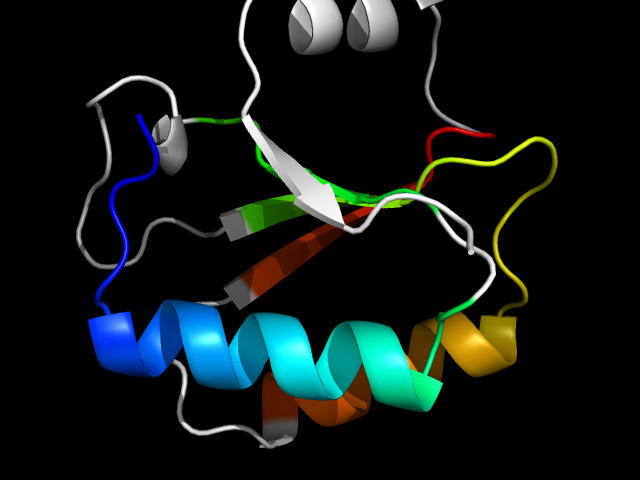 |
d1no5a_ | A6-A40,A41-A74,A75-A105 | Alpha and beta proteins (a+b) | Nucleotidyltransferase | Nucleotidyltransferase | Catalytic subunit of bi-partite nucleotidyltransferase | Hypothetical protein HI0073
|
Justification for analogy
Compare d1vaoa2 and d2mbr_1 (same superfamily, the other domain is the same), one strand and one helix are insertion. Hybrid motif in 1vao.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1vao_Ah efrpltlPPKLSLSDFNEFIQDIIRIVGseNVEVIS------------aSAIVAPRNVADVQSIVGLANKFSf------GSVVLDM---------------------
1no5_Ac -------QLDIKSEELAIVKTILQQLVPdyTVWAFGsrvkgkakkysdlDLAIISEEPLDFLARDRLKEAFSesdlpwrVDLLDWAttsedfreiirkvyvviqeke
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1vao_Ah efrpltlppKLSLSDFNEFIQDIIRIVGSENVEVI-----------sASAIVA----pRNVADVQSIVGLANKFSF-GSVVLDM-----------------------
1no5_Ac -------qlDIKSEELAIVKTILQQLVPDYTVWAFgsrvkgkakkysDLDLAIiseepLDFLARDRLKEAFSESDLpWRVDLLDwattsedfreiirkvyvviqeke
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1vao_Ah EFRPLTLPPKLSLSDFNEFIQDIIRIVGSENVEVISA--------------SAIVA---PRNVADVQSIVGLAN-K-F-SFGSVVLD-M---------------------
1no5_Ac ----QLD---IKSEELAIVKTILQQLVP-DYTVWA-FGSRVKGKAKKYSDLDLAIISEEPLDFLARDRLKEAFSESDLPWR-VDLLDWATTSEDFREIIRKVYVVIQEKE
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
1vao_Ah EFRPLTLPP--KLSLSDFNEFIQDIIRIVGSENVEVISAS---------------------AIVAPR-----NVADVQSIVGLANKFSFG-----SVVLDM------------------------
1no5_Ac ---------QLDIKSEELAIVKTILQQLV-----------PDYTVWAFGSRVKGKAKKYSDLDLAIISEEPLDFLARDRLKEAFSE----SDLPWRVDLL-DWATTSEDFREIIRKVYVVIQEKE

